
Saccharibacillus brassicae
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Paenibacillaceae; Saccharibacillus
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
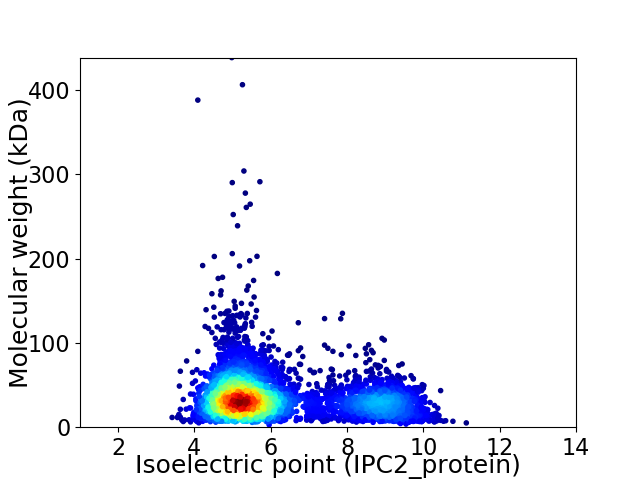
Virtual 2D-PAGE plot for 4543 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
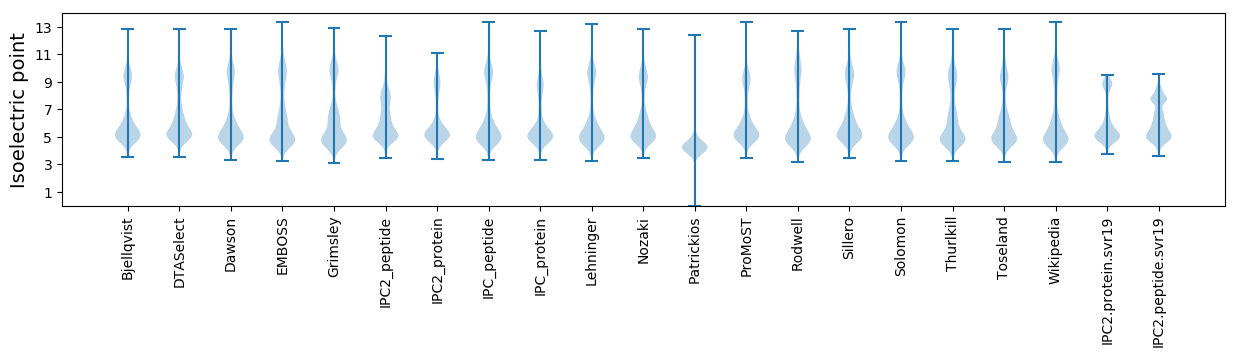
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Y6UZP1|A0A4Y6UZP1_9BACL 3-oxoacyl-[acyl-carrier-protein] synthase 2 OS=Saccharibacillus brassicae OX=2583377 GN=fabF PE=3 SV=1
MM1 pKa = 8.14RR2 pKa = 11.84LRR4 pKa = 11.84IIAGCLAFSVVVATGCSSVSTDD26 pKa = 2.52ADD28 pKa = 3.53VRR30 pKa = 11.84HH31 pKa = 5.95EE32 pKa = 4.13ADD34 pKa = 3.56KK35 pKa = 11.2AVQTTSDD42 pKa = 3.54QLNNLANLADD52 pKa = 3.68EE53 pKa = 5.51HH54 pKa = 8.5IGFVKK59 pKa = 10.52NGTMNNRR66 pKa = 11.84DD67 pKa = 3.95DD68 pKa = 3.8VTLGEE73 pKa = 4.2VMDD76 pKa = 5.02AFFDD80 pKa = 4.42GPTWQYY86 pKa = 11.29FSGTNDD92 pKa = 3.19EE93 pKa = 4.59TGDD96 pKa = 3.6TFDD99 pKa = 3.45VVEE102 pKa = 4.25FTGYY106 pKa = 10.57FLYY109 pKa = 10.84NEE111 pKa = 4.4KK112 pKa = 10.25SAKK115 pKa = 10.19ARR117 pKa = 11.84IQFILHH123 pKa = 6.65EE124 pKa = 5.28DD125 pKa = 3.45DD126 pKa = 3.59TFEE129 pKa = 4.38MGVGSYY135 pKa = 11.38NDD137 pKa = 3.32IDD139 pKa = 3.69QTALVLSLLMDD150 pKa = 4.08KK151 pKa = 10.84VYY153 pKa = 10.95EE154 pKa = 4.47SYY156 pKa = 11.18DD157 pKa = 3.52EE158 pKa = 3.99EE159 pKa = 4.85HH160 pKa = 6.85AVSTADD166 pKa = 3.29QAQAGGEE173 pKa = 4.2PVTADD178 pKa = 3.06SSQIQAAKK186 pKa = 10.75AEE188 pKa = 4.88GIGLEE193 pKa = 4.44NFYY196 pKa = 10.93KK197 pKa = 8.54WTPGEE202 pKa = 3.94NDD204 pKa = 2.93RR205 pKa = 11.84SFPLMLDD212 pKa = 3.29GEE214 pKa = 4.63EE215 pKa = 4.07IEE217 pKa = 4.76FVVGRR222 pKa = 11.84NTPNGIKK229 pKa = 8.93MQVFSSSMEE238 pKa = 4.01QGWQLPTEE246 pKa = 4.39LADD249 pKa = 4.18GSIGPFNDD257 pKa = 3.3YY258 pKa = 11.31GDD260 pKa = 4.78LIEE263 pKa = 5.62GFSLYY268 pKa = 10.54VKK270 pKa = 10.05EE271 pKa = 4.48YY272 pKa = 10.83DD273 pKa = 3.62FASDD277 pKa = 3.92GVPEE281 pKa = 4.34VVLVASDD288 pKa = 4.0GMLEE292 pKa = 4.22SYY294 pKa = 10.29VWVYY298 pKa = 10.87NYY300 pKa = 10.78NYY302 pKa = 9.37TFSEE306 pKa = 4.47YY307 pKa = 10.53DD308 pKa = 3.37VSPLEE313 pKa = 3.97LVWYY317 pKa = 10.57GEE319 pKa = 4.41GQSDD323 pKa = 3.96VQLEE327 pKa = 4.08GDD329 pKa = 4.54RR330 pKa = 11.84IVLPYY335 pKa = 10.41GSQGLYY341 pKa = 10.36EE342 pKa = 4.14EE343 pKa = 4.57YY344 pKa = 10.64VYY346 pKa = 11.29KK347 pKa = 10.64NEE349 pKa = 5.37KK350 pKa = 9.91FIQQ353 pKa = 3.6
MM1 pKa = 8.14RR2 pKa = 11.84LRR4 pKa = 11.84IIAGCLAFSVVVATGCSSVSTDD26 pKa = 2.52ADD28 pKa = 3.53VRR30 pKa = 11.84HH31 pKa = 5.95EE32 pKa = 4.13ADD34 pKa = 3.56KK35 pKa = 11.2AVQTTSDD42 pKa = 3.54QLNNLANLADD52 pKa = 3.68EE53 pKa = 5.51HH54 pKa = 8.5IGFVKK59 pKa = 10.52NGTMNNRR66 pKa = 11.84DD67 pKa = 3.95DD68 pKa = 3.8VTLGEE73 pKa = 4.2VMDD76 pKa = 5.02AFFDD80 pKa = 4.42GPTWQYY86 pKa = 11.29FSGTNDD92 pKa = 3.19EE93 pKa = 4.59TGDD96 pKa = 3.6TFDD99 pKa = 3.45VVEE102 pKa = 4.25FTGYY106 pKa = 10.57FLYY109 pKa = 10.84NEE111 pKa = 4.4KK112 pKa = 10.25SAKK115 pKa = 10.19ARR117 pKa = 11.84IQFILHH123 pKa = 6.65EE124 pKa = 5.28DD125 pKa = 3.45DD126 pKa = 3.59TFEE129 pKa = 4.38MGVGSYY135 pKa = 11.38NDD137 pKa = 3.32IDD139 pKa = 3.69QTALVLSLLMDD150 pKa = 4.08KK151 pKa = 10.84VYY153 pKa = 10.95EE154 pKa = 4.47SYY156 pKa = 11.18DD157 pKa = 3.52EE158 pKa = 3.99EE159 pKa = 4.85HH160 pKa = 6.85AVSTADD166 pKa = 3.29QAQAGGEE173 pKa = 4.2PVTADD178 pKa = 3.06SSQIQAAKK186 pKa = 10.75AEE188 pKa = 4.88GIGLEE193 pKa = 4.44NFYY196 pKa = 10.93KK197 pKa = 8.54WTPGEE202 pKa = 3.94NDD204 pKa = 2.93RR205 pKa = 11.84SFPLMLDD212 pKa = 3.29GEE214 pKa = 4.63EE215 pKa = 4.07IEE217 pKa = 4.76FVVGRR222 pKa = 11.84NTPNGIKK229 pKa = 8.93MQVFSSSMEE238 pKa = 4.01QGWQLPTEE246 pKa = 4.39LADD249 pKa = 4.18GSIGPFNDD257 pKa = 3.3YY258 pKa = 11.31GDD260 pKa = 4.78LIEE263 pKa = 5.62GFSLYY268 pKa = 10.54VKK270 pKa = 10.05EE271 pKa = 4.48YY272 pKa = 10.83DD273 pKa = 3.62FASDD277 pKa = 3.92GVPEE281 pKa = 4.34VVLVASDD288 pKa = 4.0GMLEE292 pKa = 4.22SYY294 pKa = 10.29VWVYY298 pKa = 10.87NYY300 pKa = 10.78NYY302 pKa = 9.37TFSEE306 pKa = 4.47YY307 pKa = 10.53DD308 pKa = 3.37VSPLEE313 pKa = 3.97LVWYY317 pKa = 10.57GEE319 pKa = 4.41GQSDD323 pKa = 3.96VQLEE327 pKa = 4.08GDD329 pKa = 4.54RR330 pKa = 11.84IVLPYY335 pKa = 10.41GSQGLYY341 pKa = 10.36EE342 pKa = 4.14EE343 pKa = 4.57YY344 pKa = 10.64VYY346 pKa = 11.29KK347 pKa = 10.64NEE349 pKa = 5.37KK350 pKa = 9.91FIQQ353 pKa = 3.6
Molecular weight: 39.32 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y6UW26|A0A4Y6UW26_9BACL Uncharacterized protein OS=Saccharibacillus brassicae OX=2583377 GN=FFV09_04625 PE=4 SV=1
MM1 pKa = 7.61GPTFKK6 pKa = 10.88PNTRR10 pKa = 11.84KK11 pKa = 9.83RR12 pKa = 11.84KK13 pKa = 8.74KK14 pKa = 8.74VHH16 pKa = 5.61GFRR19 pKa = 11.84KK20 pKa = 10.01RR21 pKa = 11.84MSTKK25 pKa = 10.18NGRR28 pKa = 11.84KK29 pKa = 8.87VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.2GRR39 pKa = 11.84KK40 pKa = 8.79VLSAA44 pKa = 4.05
MM1 pKa = 7.61GPTFKK6 pKa = 10.88PNTRR10 pKa = 11.84KK11 pKa = 9.83RR12 pKa = 11.84KK13 pKa = 8.74KK14 pKa = 8.74VHH16 pKa = 5.61GFRR19 pKa = 11.84KK20 pKa = 10.01RR21 pKa = 11.84MSTKK25 pKa = 10.18NGRR28 pKa = 11.84KK29 pKa = 8.87VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.2GRR39 pKa = 11.84KK40 pKa = 8.79VLSAA44 pKa = 4.05
Molecular weight: 5.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1557127 |
26 |
3974 |
342.8 |
37.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.924 ± 0.049 | 0.677 ± 0.009 |
5.429 ± 0.025 | 7.204 ± 0.044 |
3.867 ± 0.025 | 8.206 ± 0.038 |
1.928 ± 0.016 | 5.566 ± 0.032 |
4.291 ± 0.039 | 10.217 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.535 ± 0.019 | 3.204 ± 0.024 |
4.4 ± 0.028 | 3.545 ± 0.023 |
6.274 ± 0.041 | 6.141 ± 0.031 |
5.278 ± 0.037 | 6.9 ± 0.034 |
1.178 ± 0.015 | 3.236 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
