
Lake Sarah-associated circular virus-36
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 8.01
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A126GAC0|A0A126GAC0_9VIRU ATP-dependent helicase Rep OS=Lake Sarah-associated circular virus-36 OX=1685764 PE=3 SV=1
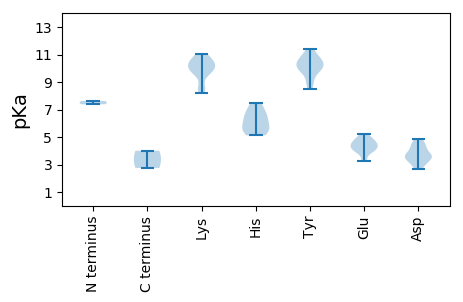
MM1 pKa = 7.38PTPKK5 pKa = 10.05KK6 pKa = 10.53SYY8 pKa = 10.67CFTLNNYY15 pKa = 8.49TDD17 pKa = 4.43GEE19 pKa = 4.43LEE21 pKa = 4.18LLRR24 pKa = 11.84VVCSEE29 pKa = 3.89EE30 pKa = 3.35ASYY33 pKa = 11.41AVIGRR38 pKa = 11.84EE39 pKa = 4.01VGEE42 pKa = 4.37TGTPHH47 pKa = 5.45LQGYY51 pKa = 9.6IRR53 pKa = 11.84FKK55 pKa = 9.77KK56 pKa = 10.08AYY58 pKa = 9.06RR59 pKa = 11.84FSTIKK64 pKa = 10.42DD65 pKa = 3.23RR66 pKa = 11.84YY67 pKa = 10.06LLRR70 pKa = 11.84CHH72 pKa = 7.05IEE74 pKa = 4.14VAAGDD79 pKa = 3.74ADD81 pKa = 4.44SNFRR85 pKa = 11.84YY86 pKa = 9.72CSKK89 pKa = 11.02DD90 pKa = 2.74GDD92 pKa = 3.58FAEE95 pKa = 5.02FGEE98 pKa = 4.89RR99 pKa = 11.84PTSGKK104 pKa = 9.62SASRR108 pKa = 11.84ADD110 pKa = 3.7LARR113 pKa = 11.84SFVSYY118 pKa = 10.39MDD120 pKa = 4.47DD121 pKa = 3.25GRR123 pKa = 11.84SGMVRR128 pKa = 11.84FAHH131 pKa = 6.25EE132 pKa = 4.84NPGTWYY138 pKa = 10.65FSGATMLRR146 pKa = 11.84NYY148 pKa = 10.12YY149 pKa = 10.39ALQPAVDD156 pKa = 3.9RR157 pKa = 11.84SDD159 pKa = 4.88VSVTWIFGDD168 pKa = 3.46PGVGKK173 pKa = 9.98SRR175 pKa = 11.84KK176 pKa = 8.74AHH178 pKa = 5.13EE179 pKa = 4.71TLPQAYY185 pKa = 10.01VKK187 pKa = 10.57DD188 pKa = 4.24PRR190 pKa = 11.84TKK192 pKa = 8.57WWNEE196 pKa = 3.29YY197 pKa = 10.75ALEE200 pKa = 4.48KK201 pKa = 10.92DD202 pKa = 4.18VIIDD206 pKa = 3.57DD207 pKa = 4.82FGPGGIDD214 pKa = 3.15INHH217 pKa = 7.45LLRR220 pKa = 11.84WFDD223 pKa = 3.45RR224 pKa = 11.84YY225 pKa = 10.74KK226 pKa = 11.0CLVEE230 pKa = 4.61SKK232 pKa = 11.01GGMIPLYY239 pKa = 9.99AVNFIVTSNFHH250 pKa = 5.93PEE252 pKa = 3.92KK253 pKa = 10.42VFSFAGDD260 pKa = 3.69PNVQLPALMRR270 pKa = 11.84RR271 pKa = 11.84MIIKK275 pKa = 10.31EE276 pKa = 4.02MVV278 pKa = 2.75
MM1 pKa = 7.38PTPKK5 pKa = 10.05KK6 pKa = 10.53SYY8 pKa = 10.67CFTLNNYY15 pKa = 8.49TDD17 pKa = 4.43GEE19 pKa = 4.43LEE21 pKa = 4.18LLRR24 pKa = 11.84VVCSEE29 pKa = 3.89EE30 pKa = 3.35ASYY33 pKa = 11.41AVIGRR38 pKa = 11.84EE39 pKa = 4.01VGEE42 pKa = 4.37TGTPHH47 pKa = 5.45LQGYY51 pKa = 9.6IRR53 pKa = 11.84FKK55 pKa = 9.77KK56 pKa = 10.08AYY58 pKa = 9.06RR59 pKa = 11.84FSTIKK64 pKa = 10.42DD65 pKa = 3.23RR66 pKa = 11.84YY67 pKa = 10.06LLRR70 pKa = 11.84CHH72 pKa = 7.05IEE74 pKa = 4.14VAAGDD79 pKa = 3.74ADD81 pKa = 4.44SNFRR85 pKa = 11.84YY86 pKa = 9.72CSKK89 pKa = 11.02DD90 pKa = 2.74GDD92 pKa = 3.58FAEE95 pKa = 5.02FGEE98 pKa = 4.89RR99 pKa = 11.84PTSGKK104 pKa = 9.62SASRR108 pKa = 11.84ADD110 pKa = 3.7LARR113 pKa = 11.84SFVSYY118 pKa = 10.39MDD120 pKa = 4.47DD121 pKa = 3.25GRR123 pKa = 11.84SGMVRR128 pKa = 11.84FAHH131 pKa = 6.25EE132 pKa = 4.84NPGTWYY138 pKa = 10.65FSGATMLRR146 pKa = 11.84NYY148 pKa = 10.12YY149 pKa = 10.39ALQPAVDD156 pKa = 3.9RR157 pKa = 11.84SDD159 pKa = 4.88VSVTWIFGDD168 pKa = 3.46PGVGKK173 pKa = 9.98SRR175 pKa = 11.84KK176 pKa = 8.74AHH178 pKa = 5.13EE179 pKa = 4.71TLPQAYY185 pKa = 10.01VKK187 pKa = 10.57DD188 pKa = 4.24PRR190 pKa = 11.84TKK192 pKa = 8.57WWNEE196 pKa = 3.29YY197 pKa = 10.75ALEE200 pKa = 4.48KK201 pKa = 10.92DD202 pKa = 4.18VIIDD206 pKa = 3.57DD207 pKa = 4.82FGPGGIDD214 pKa = 3.15INHH217 pKa = 7.45LLRR220 pKa = 11.84WFDD223 pKa = 3.45RR224 pKa = 11.84YY225 pKa = 10.74KK226 pKa = 11.0CLVEE230 pKa = 4.61SKK232 pKa = 11.01GGMIPLYY239 pKa = 9.99AVNFIVTSNFHH250 pKa = 5.93PEE252 pKa = 3.92KK253 pKa = 10.42VFSFAGDD260 pKa = 3.69PNVQLPALMRR270 pKa = 11.84RR271 pKa = 11.84MIIKK275 pKa = 10.31EE276 pKa = 4.02MVV278 pKa = 2.75
Molecular weight: 31.62 kDa
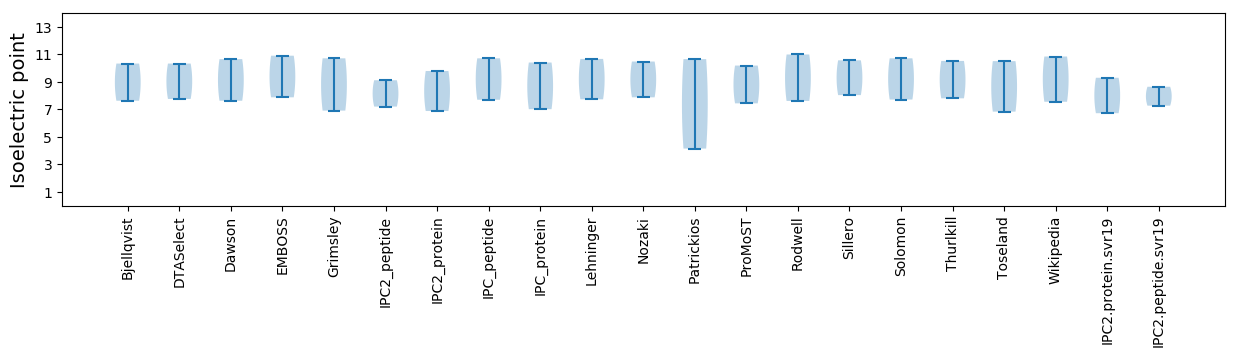
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A126GAC0|A0A126GAC0_9VIRU ATP-dependent helicase Rep OS=Lake Sarah-associated circular virus-36 OX=1685764 PE=3 SV=1
MM1 pKa = 7.62AMTGRR6 pKa = 11.84KK7 pKa = 8.47RR8 pKa = 11.84VYY10 pKa = 10.27GGKK13 pKa = 8.17PCKK16 pKa = 10.15NRR18 pKa = 11.84FIKK21 pKa = 9.64KK22 pKa = 8.43RR23 pKa = 11.84RR24 pKa = 11.84MMARR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84SKK33 pKa = 10.63KK34 pKa = 10.22AVGLSTRR41 pKa = 11.84TLAPTDD47 pKa = 3.69FHH49 pKa = 6.63TKK51 pKa = 9.7GRR53 pKa = 11.84MLKK56 pKa = 9.72KK57 pKa = 10.04RR58 pKa = 11.84QWRR61 pKa = 11.84NLLWRR66 pKa = 11.84NTLTSEE72 pKa = 4.48KK73 pKa = 10.16YY74 pKa = 10.18RR75 pKa = 11.84SILMATTGLSTPGNHH90 pKa = 5.45TTANWSDD97 pKa = 3.93LEE99 pKa = 4.31IMSNTQPFWTSAGGMQPVNQGSGVPTLAPDD129 pKa = 4.2HH130 pKa = 6.47IIIRR134 pKa = 11.84GGTFYY139 pKa = 11.16FSMINPLGNTTDD151 pKa = 2.82IRR153 pKa = 11.84VRR155 pKa = 11.84LQLLYY160 pKa = 9.84PKK162 pKa = 10.6QNITRR167 pKa = 11.84YY168 pKa = 9.88ASLVRR173 pKa = 11.84TNVPLIDD180 pKa = 4.63WITSLGGNQPTIWRR194 pKa = 11.84MQDD197 pKa = 2.71AGDD200 pKa = 3.77YY201 pKa = 8.79NQYY204 pKa = 10.65YY205 pKa = 9.18YY206 pKa = 11.29NPVMEE211 pKa = 5.24KK212 pKa = 10.48EE213 pKa = 4.6IILKK217 pKa = 10.26PGDD220 pKa = 3.53SFEE223 pKa = 4.92EE224 pKa = 4.03YY225 pKa = 10.68RR226 pKa = 11.84KK227 pKa = 9.74MKK229 pKa = 9.59VQRR232 pKa = 11.84LDD234 pKa = 3.15TEE236 pKa = 4.36AFLNGAAGFPSWLVYY251 pKa = 10.21ISNLDD256 pKa = 3.24GAAVQNVTVVSGHH269 pKa = 5.55SLSFALADD277 pKa = 3.52VV278 pKa = 4.0
MM1 pKa = 7.62AMTGRR6 pKa = 11.84KK7 pKa = 8.47RR8 pKa = 11.84VYY10 pKa = 10.27GGKK13 pKa = 8.17PCKK16 pKa = 10.15NRR18 pKa = 11.84FIKK21 pKa = 9.64KK22 pKa = 8.43RR23 pKa = 11.84RR24 pKa = 11.84MMARR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84SKK33 pKa = 10.63KK34 pKa = 10.22AVGLSTRR41 pKa = 11.84TLAPTDD47 pKa = 3.69FHH49 pKa = 6.63TKK51 pKa = 9.7GRR53 pKa = 11.84MLKK56 pKa = 9.72KK57 pKa = 10.04RR58 pKa = 11.84QWRR61 pKa = 11.84NLLWRR66 pKa = 11.84NTLTSEE72 pKa = 4.48KK73 pKa = 10.16YY74 pKa = 10.18RR75 pKa = 11.84SILMATTGLSTPGNHH90 pKa = 5.45TTANWSDD97 pKa = 3.93LEE99 pKa = 4.31IMSNTQPFWTSAGGMQPVNQGSGVPTLAPDD129 pKa = 4.2HH130 pKa = 6.47IIIRR134 pKa = 11.84GGTFYY139 pKa = 11.16FSMINPLGNTTDD151 pKa = 2.82IRR153 pKa = 11.84VRR155 pKa = 11.84LQLLYY160 pKa = 9.84PKK162 pKa = 10.6QNITRR167 pKa = 11.84YY168 pKa = 9.88ASLVRR173 pKa = 11.84TNVPLIDD180 pKa = 4.63WITSLGGNQPTIWRR194 pKa = 11.84MQDD197 pKa = 2.71AGDD200 pKa = 3.77YY201 pKa = 8.79NQYY204 pKa = 10.65YY205 pKa = 9.18YY206 pKa = 11.29NPVMEE211 pKa = 5.24KK212 pKa = 10.48EE213 pKa = 4.6IILKK217 pKa = 10.26PGDD220 pKa = 3.53SFEE223 pKa = 4.92EE224 pKa = 4.03YY225 pKa = 10.68RR226 pKa = 11.84KK227 pKa = 9.74MKK229 pKa = 9.59VQRR232 pKa = 11.84LDD234 pKa = 3.15TEE236 pKa = 4.36AFLNGAAGFPSWLVYY251 pKa = 10.21ISNLDD256 pKa = 3.24GAAVQNVTVVSGHH269 pKa = 5.55SLSFALADD277 pKa = 3.52VV278 pKa = 4.0
Molecular weight: 31.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
556 |
278 |
278 |
278.0 |
31.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.655 ± 0.332 | 1.079 ± 0.443 |
5.396 ± 0.886 | 4.137 ± 0.997 |
4.496 ± 0.776 | 7.914 ± 0.0 |
1.799 ± 0.222 | 5.036 ± 0.222 |
5.755 ± 0.0 | 7.554 ± 0.665 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.597 ± 0.443 | 4.856 ± 0.776 |
5.036 ± 0.0 | 2.698 ± 0.776 |
7.734 ± 0.332 | 6.655 ± 0.111 |
6.655 ± 1.219 | 6.115 ± 0.443 |
2.158 ± 0.222 | 4.676 ± 0.443 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
