
alpha proteobacterium AAP81b
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; unclassified Alphaproteobacteria
Average proteome isoelectric point is 6.74
Get precalculated fractions of proteins
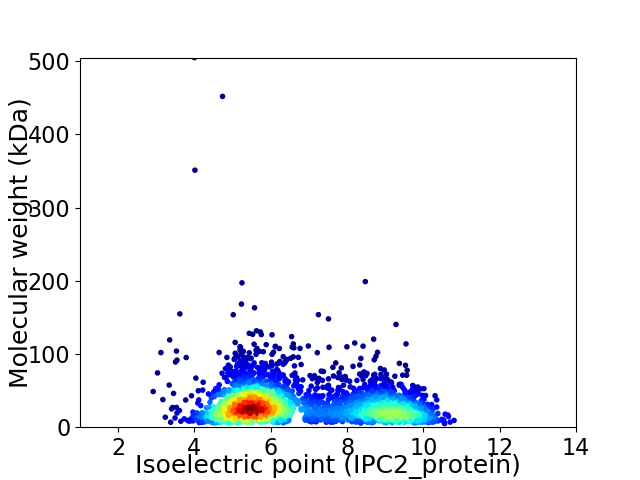
Virtual 2D-PAGE plot for 3055 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0N0JUJ6|A0A0N0JUJ6_9PROT Peptidase_M56 domain-containing protein (Fragment) OS=alpha proteobacterium AAP81b OX=1523432 GN=IP88_11860 PE=4 SV=1
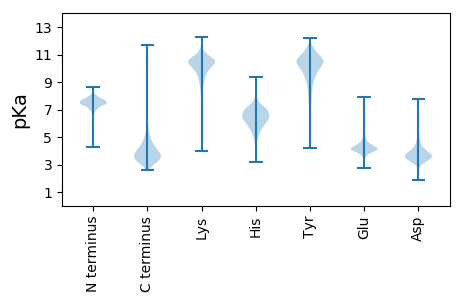
MM1 pKa = 7.55GAQSDD6 pKa = 4.09PNKK9 pKa = 10.23EE10 pKa = 3.68VLYY13 pKa = 11.11YY14 pKa = 10.96GDD16 pKa = 3.7TGEE19 pKa = 5.43LIGTEE24 pKa = 4.34SFSLDD29 pKa = 3.35PFAGGSYY36 pKa = 9.85LGKK39 pKa = 9.77PIADD43 pKa = 3.43LEE45 pKa = 4.5FVTEE49 pKa = 3.91NLVRR53 pKa = 11.84SGSNWLNGANYY64 pKa = 10.08GVLDD68 pKa = 4.75DD69 pKa = 4.7GVLNFGFWNNIQEE82 pKa = 4.47LQNSYY87 pKa = 10.56YY88 pKa = 11.18VNDD91 pKa = 4.48DD92 pKa = 3.03GSIRR96 pKa = 11.84FSEE99 pKa = 4.4ANPATFSAFSTAQRR113 pKa = 11.84AVARR117 pKa = 11.84TTIALWDD124 pKa = 4.06DD125 pKa = 4.81LIDD128 pKa = 3.35ISFRR132 pKa = 11.84EE133 pKa = 4.26TRR135 pKa = 11.84SGVADD140 pKa = 3.11ITYY143 pKa = 10.62GNTAAGANVQAYY155 pKa = 9.85AYY157 pKa = 10.43LPFGDD162 pKa = 5.07VYY164 pKa = 11.54DD165 pKa = 4.85DD166 pKa = 4.59YY167 pKa = 11.54YY168 pKa = 11.83AQFGWEE174 pKa = 3.84TAQIGRR180 pKa = 11.84LGGDD184 pKa = 2.95VWIGGNVGSNFTPLQNSYY202 pKa = 11.05YY203 pKa = 10.86SVITQIHH210 pKa = 4.92EE211 pKa = 4.06TGHH214 pKa = 6.61ALGLSHH220 pKa = 7.5PGDD223 pKa = 4.15YY224 pKa = 10.47NASDD228 pKa = 4.72DD229 pKa = 3.9NDD231 pKa = 5.33GIPGPDD237 pKa = 4.49PITYY241 pKa = 10.03TNDD244 pKa = 2.64AYY246 pKa = 10.67FAQDD250 pKa = 3.09SLQYY254 pKa = 11.3SVMSYY259 pKa = 10.38FDD261 pKa = 3.77AYY263 pKa = 9.06EE264 pKa = 4.12TGAQHH269 pKa = 7.46IDD271 pKa = 2.78WTLTNFAYY279 pKa = 10.31AATPLVHH286 pKa = 7.34DD287 pKa = 4.51VAAIQALYY295 pKa = 10.59GVDD298 pKa = 3.05TTTRR302 pKa = 11.84TGDD305 pKa = 3.16TVYY308 pKa = 11.03GFNSTANRR316 pKa = 11.84TAYY319 pKa = 10.46DD320 pKa = 3.59FTSNTRR326 pKa = 11.84PIVTIWDD333 pKa = 3.32AGGIDD338 pKa = 4.15TIDD341 pKa = 3.43FSGWNTPSVIDD352 pKa = 4.18LNEE355 pKa = 4.09GAFSSGGGTEE365 pKa = 3.93QFLTLAQVNANRR377 pKa = 11.84AALGFAPRR385 pKa = 11.84SQATFNFYY393 pKa = 10.86EE394 pKa = 4.36GLKK397 pKa = 10.35AQLGLTNGLFKK408 pKa = 11.24DD409 pKa = 4.03NVSIAYY415 pKa = 8.71GATIEE420 pKa = 4.11NATGGGGNDD429 pKa = 2.82RR430 pKa = 11.84LIANNVANVLTGNGGVDD447 pKa = 4.1FASYY451 pKa = 9.88EE452 pKa = 3.94AATAGVTVNLATGGTAGYY470 pKa = 9.93AAGDD474 pKa = 3.86SYY476 pKa = 11.99VGIEE480 pKa = 4.14GVIGSDD486 pKa = 3.96FNDD489 pKa = 3.53VLDD492 pKa = 5.03GSAGDD497 pKa = 4.42DD498 pKa = 3.55LFAVSGGADD507 pKa = 4.25AINGGAGSDD516 pKa = 3.19TLTFQFAEE524 pKa = 4.13AGVNVSLTGTGLGGHH539 pKa = 6.25TLTSIEE545 pKa = 4.47NLTGSAFNDD554 pKa = 3.71TLGGDD559 pKa = 3.53SAANVINGGAGIDD572 pKa = 3.45TLFYY576 pKa = 10.94AGATSGVTVNLATGGTRR593 pKa = 11.84GLAAGDD599 pKa = 3.73TYY601 pKa = 11.76VSIEE605 pKa = 4.65NITATNFADD614 pKa = 4.41NLTGDD619 pKa = 3.48AGVNVIQGGLGADD632 pKa = 3.82TLTGGAGDD640 pKa = 3.78VLSYY644 pKa = 11.14ADD646 pKa = 3.46SAAAITINLGNNYY659 pKa = 9.95AAGGTAARR667 pKa = 11.84DD668 pKa = 3.83VISGFDD674 pKa = 3.42NVTASAFNDD683 pKa = 3.99NITGDD688 pKa = 3.59NDD690 pKa = 3.99ANTLTGGDD698 pKa = 4.17GNDD701 pKa = 3.75TLIGAAGADD710 pKa = 3.48QLFGGIGNDD719 pKa = 4.15SIRR722 pKa = 11.84AGNDD726 pKa = 2.42NDD728 pKa = 4.07YY729 pKa = 11.35IEE731 pKa = 5.33AGNGNDD737 pKa = 3.7TVLGDD742 pKa = 4.06AQNDD746 pKa = 4.12TLFGGAGDD754 pKa = 4.66DD755 pKa = 3.86NLNGGGNDD763 pKa = 3.79DD764 pKa = 4.28YY765 pKa = 11.9LNGGTGTNTLTGGTGLDD782 pKa = 3.11IFDD785 pKa = 5.29FDD787 pKa = 4.19TASFTDD793 pKa = 4.43TITDD797 pKa = 3.94FRR799 pKa = 11.84TGLDD803 pKa = 3.86KK804 pKa = 10.74IDD806 pKa = 3.65VSDD809 pKa = 3.31IAGFTFIGGAAFSGANQLRR828 pKa = 11.84FYY830 pKa = 10.9TDD832 pKa = 3.1GANRR836 pKa = 11.84ALAGDD841 pKa = 3.54IDD843 pKa = 4.67GDD845 pKa = 4.28GVADD849 pKa = 4.48LVIHH853 pKa = 5.92LTGAVSVVAGDD864 pKa = 4.79LILVV868 pKa = 4.17
MM1 pKa = 7.55GAQSDD6 pKa = 4.09PNKK9 pKa = 10.23EE10 pKa = 3.68VLYY13 pKa = 11.11YY14 pKa = 10.96GDD16 pKa = 3.7TGEE19 pKa = 5.43LIGTEE24 pKa = 4.34SFSLDD29 pKa = 3.35PFAGGSYY36 pKa = 9.85LGKK39 pKa = 9.77PIADD43 pKa = 3.43LEE45 pKa = 4.5FVTEE49 pKa = 3.91NLVRR53 pKa = 11.84SGSNWLNGANYY64 pKa = 10.08GVLDD68 pKa = 4.75DD69 pKa = 4.7GVLNFGFWNNIQEE82 pKa = 4.47LQNSYY87 pKa = 10.56YY88 pKa = 11.18VNDD91 pKa = 4.48DD92 pKa = 3.03GSIRR96 pKa = 11.84FSEE99 pKa = 4.4ANPATFSAFSTAQRR113 pKa = 11.84AVARR117 pKa = 11.84TTIALWDD124 pKa = 4.06DD125 pKa = 4.81LIDD128 pKa = 3.35ISFRR132 pKa = 11.84EE133 pKa = 4.26TRR135 pKa = 11.84SGVADD140 pKa = 3.11ITYY143 pKa = 10.62GNTAAGANVQAYY155 pKa = 9.85AYY157 pKa = 10.43LPFGDD162 pKa = 5.07VYY164 pKa = 11.54DD165 pKa = 4.85DD166 pKa = 4.59YY167 pKa = 11.54YY168 pKa = 11.83AQFGWEE174 pKa = 3.84TAQIGRR180 pKa = 11.84LGGDD184 pKa = 2.95VWIGGNVGSNFTPLQNSYY202 pKa = 11.05YY203 pKa = 10.86SVITQIHH210 pKa = 4.92EE211 pKa = 4.06TGHH214 pKa = 6.61ALGLSHH220 pKa = 7.5PGDD223 pKa = 4.15YY224 pKa = 10.47NASDD228 pKa = 4.72DD229 pKa = 3.9NDD231 pKa = 5.33GIPGPDD237 pKa = 4.49PITYY241 pKa = 10.03TNDD244 pKa = 2.64AYY246 pKa = 10.67FAQDD250 pKa = 3.09SLQYY254 pKa = 11.3SVMSYY259 pKa = 10.38FDD261 pKa = 3.77AYY263 pKa = 9.06EE264 pKa = 4.12TGAQHH269 pKa = 7.46IDD271 pKa = 2.78WTLTNFAYY279 pKa = 10.31AATPLVHH286 pKa = 7.34DD287 pKa = 4.51VAAIQALYY295 pKa = 10.59GVDD298 pKa = 3.05TTTRR302 pKa = 11.84TGDD305 pKa = 3.16TVYY308 pKa = 11.03GFNSTANRR316 pKa = 11.84TAYY319 pKa = 10.46DD320 pKa = 3.59FTSNTRR326 pKa = 11.84PIVTIWDD333 pKa = 3.32AGGIDD338 pKa = 4.15TIDD341 pKa = 3.43FSGWNTPSVIDD352 pKa = 4.18LNEE355 pKa = 4.09GAFSSGGGTEE365 pKa = 3.93QFLTLAQVNANRR377 pKa = 11.84AALGFAPRR385 pKa = 11.84SQATFNFYY393 pKa = 10.86EE394 pKa = 4.36GLKK397 pKa = 10.35AQLGLTNGLFKK408 pKa = 11.24DD409 pKa = 4.03NVSIAYY415 pKa = 8.71GATIEE420 pKa = 4.11NATGGGGNDD429 pKa = 2.82RR430 pKa = 11.84LIANNVANVLTGNGGVDD447 pKa = 4.1FASYY451 pKa = 9.88EE452 pKa = 3.94AATAGVTVNLATGGTAGYY470 pKa = 9.93AAGDD474 pKa = 3.86SYY476 pKa = 11.99VGIEE480 pKa = 4.14GVIGSDD486 pKa = 3.96FNDD489 pKa = 3.53VLDD492 pKa = 5.03GSAGDD497 pKa = 4.42DD498 pKa = 3.55LFAVSGGADD507 pKa = 4.25AINGGAGSDD516 pKa = 3.19TLTFQFAEE524 pKa = 4.13AGVNVSLTGTGLGGHH539 pKa = 6.25TLTSIEE545 pKa = 4.47NLTGSAFNDD554 pKa = 3.71TLGGDD559 pKa = 3.53SAANVINGGAGIDD572 pKa = 3.45TLFYY576 pKa = 10.94AGATSGVTVNLATGGTRR593 pKa = 11.84GLAAGDD599 pKa = 3.73TYY601 pKa = 11.76VSIEE605 pKa = 4.65NITATNFADD614 pKa = 4.41NLTGDD619 pKa = 3.48AGVNVIQGGLGADD632 pKa = 3.82TLTGGAGDD640 pKa = 3.78VLSYY644 pKa = 11.14ADD646 pKa = 3.46SAAAITINLGNNYY659 pKa = 9.95AAGGTAARR667 pKa = 11.84DD668 pKa = 3.83VISGFDD674 pKa = 3.42NVTASAFNDD683 pKa = 3.99NITGDD688 pKa = 3.59NDD690 pKa = 3.99ANTLTGGDD698 pKa = 4.17GNDD701 pKa = 3.75TLIGAAGADD710 pKa = 3.48QLFGGIGNDD719 pKa = 4.15SIRR722 pKa = 11.84AGNDD726 pKa = 2.42NDD728 pKa = 4.07YY729 pKa = 11.35IEE731 pKa = 5.33AGNGNDD737 pKa = 3.7TVLGDD742 pKa = 4.06AQNDD746 pKa = 4.12TLFGGAGDD754 pKa = 4.66DD755 pKa = 3.86NLNGGGNDD763 pKa = 3.79DD764 pKa = 4.28YY765 pKa = 11.9LNGGTGTNTLTGGTGLDD782 pKa = 3.11IFDD785 pKa = 5.29FDD787 pKa = 4.19TASFTDD793 pKa = 4.43TITDD797 pKa = 3.94FRR799 pKa = 11.84TGLDD803 pKa = 3.86KK804 pKa = 10.74IDD806 pKa = 3.65VSDD809 pKa = 3.31IAGFTFIGGAAFSGANQLRR828 pKa = 11.84FYY830 pKa = 10.9TDD832 pKa = 3.1GANRR836 pKa = 11.84ALAGDD841 pKa = 3.54IDD843 pKa = 4.67GDD845 pKa = 4.28GVADD849 pKa = 4.48LVIHH853 pKa = 5.92LTGAVSVVAGDD864 pKa = 4.79LILVV868 pKa = 4.17
Molecular weight: 89.05 kDa
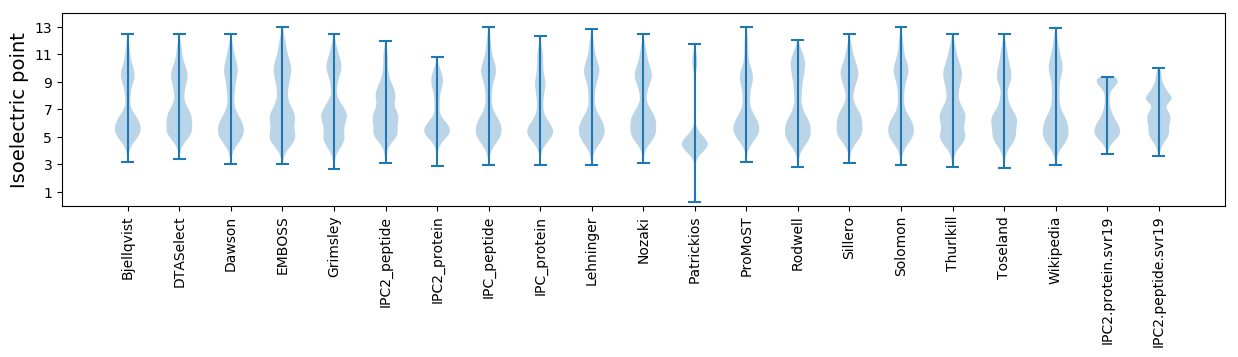
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0N0K463|A0A0N0K463_9PROT Uncharacterized protein OS=alpha proteobacterium AAP81b OX=1523432 GN=IP88_02650 PE=4 SV=1
MM1 pKa = 6.79TAAPRR6 pKa = 11.84TIGRR10 pKa = 11.84PPRR13 pKa = 11.84RR14 pKa = 11.84QARR17 pKa = 11.84GRR19 pKa = 11.84QAIAALALLCGLALAGADD37 pKa = 3.62PAAAQPSGPWKK48 pKa = 10.5GDD50 pKa = 3.01PCRR53 pKa = 11.84YY54 pKa = 9.13FRR56 pKa = 11.84QQRR59 pKa = 11.84AAGRR63 pKa = 11.84CTGVGNWKK71 pKa = 10.33VNEE74 pKa = 4.2GDD76 pKa = 4.06SFTTYY81 pKa = 10.38HH82 pKa = 7.1ADD84 pKa = 3.08GTLRR88 pKa = 11.84SNTPPPPPPPPLPPLPPPSPDD109 pKa = 2.68WVPPEE114 pKa = 4.19GFEE117 pKa = 4.29PPRR120 pKa = 11.84EE121 pKa = 4.2AAVTAPMPAATAVGAVSPGPVSSGPWRR148 pKa = 11.84GDD150 pKa = 3.02PCGHH154 pKa = 6.35FRR156 pKa = 11.84RR157 pKa = 11.84GNVSGRR163 pKa = 11.84CDD165 pKa = 3.12KK166 pKa = 11.23GVWKK170 pKa = 10.21QATGADD176 pKa = 3.33RR177 pKa = 11.84FQWFFADD184 pKa = 4.33GSSRR188 pKa = 11.84TVTSAPAVNNSKK200 pKa = 9.11TMTLTIAALPPAVRR214 pKa = 11.84VEE216 pKa = 4.07ATIAPPDD223 pKa = 4.16PPSGAWRR230 pKa = 11.84GDD232 pKa = 3.0PCGYY236 pKa = 9.73FRR238 pKa = 11.84RR239 pKa = 11.84SNAVGRR245 pKa = 11.84CQNGSWRR252 pKa = 11.84IAAGTGRR259 pKa = 11.84LTLFLADD266 pKa = 3.73GSSRR270 pKa = 11.84TITVQPALPATKK282 pKa = 10.28
MM1 pKa = 6.79TAAPRR6 pKa = 11.84TIGRR10 pKa = 11.84PPRR13 pKa = 11.84RR14 pKa = 11.84QARR17 pKa = 11.84GRR19 pKa = 11.84QAIAALALLCGLALAGADD37 pKa = 3.62PAAAQPSGPWKK48 pKa = 10.5GDD50 pKa = 3.01PCRR53 pKa = 11.84YY54 pKa = 9.13FRR56 pKa = 11.84QQRR59 pKa = 11.84AAGRR63 pKa = 11.84CTGVGNWKK71 pKa = 10.33VNEE74 pKa = 4.2GDD76 pKa = 4.06SFTTYY81 pKa = 10.38HH82 pKa = 7.1ADD84 pKa = 3.08GTLRR88 pKa = 11.84SNTPPPPPPPPLPPLPPPSPDD109 pKa = 2.68WVPPEE114 pKa = 4.19GFEE117 pKa = 4.29PPRR120 pKa = 11.84EE121 pKa = 4.2AAVTAPMPAATAVGAVSPGPVSSGPWRR148 pKa = 11.84GDD150 pKa = 3.02PCGHH154 pKa = 6.35FRR156 pKa = 11.84RR157 pKa = 11.84GNVSGRR163 pKa = 11.84CDD165 pKa = 3.12KK166 pKa = 11.23GVWKK170 pKa = 10.21QATGADD176 pKa = 3.33RR177 pKa = 11.84FQWFFADD184 pKa = 4.33GSSRR188 pKa = 11.84TVTSAPAVNNSKK200 pKa = 9.11TMTLTIAALPPAVRR214 pKa = 11.84VEE216 pKa = 4.07ATIAPPDD223 pKa = 4.16PPSGAWRR230 pKa = 11.84GDD232 pKa = 3.0PCGYY236 pKa = 9.73FRR238 pKa = 11.84RR239 pKa = 11.84SNAVGRR245 pKa = 11.84CQNGSWRR252 pKa = 11.84IAAGTGRR259 pKa = 11.84LTLFLADD266 pKa = 3.73GSSRR270 pKa = 11.84TITVQPALPATKK282 pKa = 10.28
Molecular weight: 29.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
934311 |
41 |
5206 |
305.8 |
32.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
15.746 ± 0.082 | 0.696 ± 0.014 |
5.94 ± 0.035 | 4.616 ± 0.043 |
3.496 ± 0.027 | 9.493 ± 0.074 |
1.814 ± 0.027 | 4.65 ± 0.027 |
2.391 ± 0.037 | 10.31 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.053 ± 0.026 | 2.355 ± 0.053 |
5.587 ± 0.048 | 2.615 ± 0.023 |
7.497 ± 0.057 | 4.509 ± 0.037 |
5.398 ± 0.071 | 7.453 ± 0.037 |
1.432 ± 0.022 | 1.95 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
