
Jeotgalibacillus campisalis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Planococcaceae; Jeotgalibacillus
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
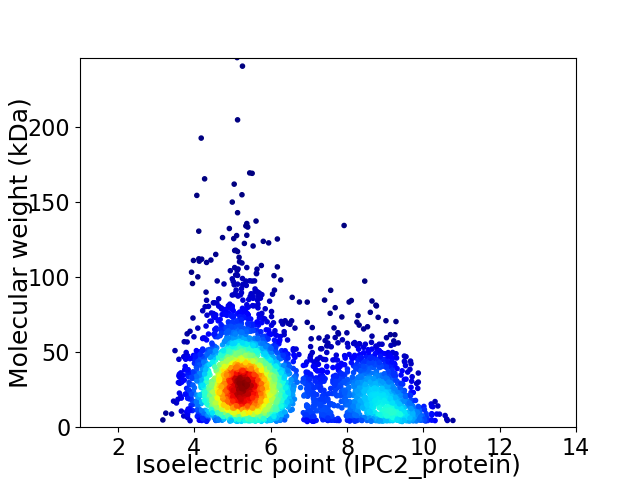
Virtual 2D-PAGE plot for 3729 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
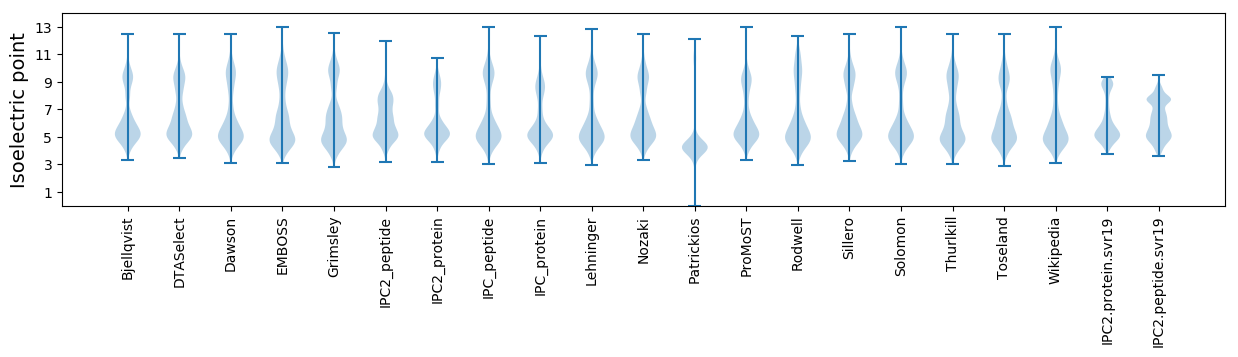
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C2R024|A0A0C2R024_9BACL Uncharacterized protein OS=Jeotgalibacillus campisalis OX=220754 GN=KR50_31850 PE=4 SV=1
MM1 pKa = 7.47LKK3 pKa = 10.13KK4 pKa = 10.56VLLIVSIVLTGAVVYY19 pKa = 9.46FGQDD23 pKa = 2.49AWKK26 pKa = 10.24DD27 pKa = 3.63SQKK30 pKa = 10.44EE31 pKa = 3.99VHH33 pKa = 6.63RR34 pKa = 11.84SSSSVVEE41 pKa = 4.25STSNANSEE49 pKa = 4.33DD50 pKa = 3.82SADD53 pKa = 3.49SSTSIEE59 pKa = 4.1NEE61 pKa = 4.22SKK63 pKa = 10.62KK64 pKa = 10.97GSADD68 pKa = 3.48EE69 pKa = 5.23LEE71 pKa = 4.9DD72 pKa = 5.55LIANQPQDD80 pKa = 3.49VQEE83 pKa = 4.12FWIEE87 pKa = 4.2SKK89 pKa = 10.99EE90 pKa = 4.24SGDD93 pKa = 3.7TVDD96 pKa = 3.05ITFVATEE103 pKa = 3.97SAVALEE109 pKa = 4.64EE110 pKa = 3.92NWTTLIEE117 pKa = 4.03EE118 pKa = 4.7SFLSSYY124 pKa = 10.77EE125 pKa = 4.28GIDD128 pKa = 3.49FAFSLITHH136 pKa = 6.19EE137 pKa = 4.76NEE139 pKa = 4.03GTSEE143 pKa = 4.03DD144 pKa = 4.2WLTSLQSEE152 pKa = 4.21AVTFEE157 pKa = 3.93GQDD160 pKa = 2.88IVLYY164 pKa = 8.32EE165 pKa = 4.25LPVINDD171 pKa = 3.13NGMLSSQDD179 pKa = 3.11QIYY182 pKa = 7.64YY183 pKa = 8.64TNRR186 pKa = 11.84FLEE189 pKa = 4.48EE190 pKa = 3.94MQSNFPEE197 pKa = 4.2THH199 pKa = 6.85LFTLPSQPLYY209 pKa = 10.99NSTYY213 pKa = 10.17YY214 pKa = 10.01PGEE217 pKa = 4.27LEE219 pKa = 4.29TVQEE223 pKa = 4.44VVEE226 pKa = 4.2EE227 pKa = 3.86QGIPFLNHH235 pKa = 5.46WEE237 pKa = 4.14DD238 pKa = 3.3WPSIDD243 pKa = 6.06DD244 pKa = 4.16EE245 pKa = 4.28EE246 pKa = 5.63LEE248 pKa = 5.33NYY250 pKa = 8.17LTDD253 pKa = 5.31DD254 pKa = 3.84NDD256 pKa = 3.95PNEE259 pKa = 4.59EE260 pKa = 4.39GNTIWGTYY268 pKa = 8.99LVDD271 pKa = 3.67YY272 pKa = 10.48FSTNN276 pKa = 2.7
MM1 pKa = 7.47LKK3 pKa = 10.13KK4 pKa = 10.56VLLIVSIVLTGAVVYY19 pKa = 9.46FGQDD23 pKa = 2.49AWKK26 pKa = 10.24DD27 pKa = 3.63SQKK30 pKa = 10.44EE31 pKa = 3.99VHH33 pKa = 6.63RR34 pKa = 11.84SSSSVVEE41 pKa = 4.25STSNANSEE49 pKa = 4.33DD50 pKa = 3.82SADD53 pKa = 3.49SSTSIEE59 pKa = 4.1NEE61 pKa = 4.22SKK63 pKa = 10.62KK64 pKa = 10.97GSADD68 pKa = 3.48EE69 pKa = 5.23LEE71 pKa = 4.9DD72 pKa = 5.55LIANQPQDD80 pKa = 3.49VQEE83 pKa = 4.12FWIEE87 pKa = 4.2SKK89 pKa = 10.99EE90 pKa = 4.24SGDD93 pKa = 3.7TVDD96 pKa = 3.05ITFVATEE103 pKa = 3.97SAVALEE109 pKa = 4.64EE110 pKa = 3.92NWTTLIEE117 pKa = 4.03EE118 pKa = 4.7SFLSSYY124 pKa = 10.77EE125 pKa = 4.28GIDD128 pKa = 3.49FAFSLITHH136 pKa = 6.19EE137 pKa = 4.76NEE139 pKa = 4.03GTSEE143 pKa = 4.03DD144 pKa = 4.2WLTSLQSEE152 pKa = 4.21AVTFEE157 pKa = 3.93GQDD160 pKa = 2.88IVLYY164 pKa = 8.32EE165 pKa = 4.25LPVINDD171 pKa = 3.13NGMLSSQDD179 pKa = 3.11QIYY182 pKa = 7.64YY183 pKa = 8.64TNRR186 pKa = 11.84FLEE189 pKa = 4.48EE190 pKa = 3.94MQSNFPEE197 pKa = 4.2THH199 pKa = 6.85LFTLPSQPLYY209 pKa = 10.99NSTYY213 pKa = 10.17YY214 pKa = 10.01PGEE217 pKa = 4.27LEE219 pKa = 4.29TVQEE223 pKa = 4.44VVEE226 pKa = 4.2EE227 pKa = 3.86QGIPFLNHH235 pKa = 5.46WEE237 pKa = 4.14DD238 pKa = 3.3WPSIDD243 pKa = 6.06DD244 pKa = 4.16EE245 pKa = 4.28EE246 pKa = 5.63LEE248 pKa = 5.33NYY250 pKa = 8.17LTDD253 pKa = 5.31DD254 pKa = 3.84NDD256 pKa = 3.95PNEE259 pKa = 4.59EE260 pKa = 4.39GNTIWGTYY268 pKa = 8.99LVDD271 pKa = 3.67YY272 pKa = 10.48FSTNN276 pKa = 2.7
Molecular weight: 31.25 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C2VPM0|A0A0C2VPM0_9BACL RNA 2' 3'-cyclic phosphodiesterase OS=Jeotgalibacillus campisalis OX=220754 GN=KR50_30610 PE=3 SV=1
MM1 pKa = 7.34LLSWVVPRR9 pKa = 11.84IKK11 pKa = 10.66ASLHH15 pKa = 4.74KK16 pKa = 10.48QGAIFMRR23 pKa = 11.84FFNLKK28 pKa = 9.91LKK30 pKa = 10.72RR31 pKa = 11.84PVQARR36 pKa = 11.84QVV38 pKa = 3.21
MM1 pKa = 7.34LLSWVVPRR9 pKa = 11.84IKK11 pKa = 10.66ASLHH15 pKa = 4.74KK16 pKa = 10.48QGAIFMRR23 pKa = 11.84FFNLKK28 pKa = 9.91LKK30 pKa = 10.72RR31 pKa = 11.84PVQARR36 pKa = 11.84QVV38 pKa = 3.21
Molecular weight: 4.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1041369 |
37 |
2211 |
279.3 |
31.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.313 ± 0.045 | 0.641 ± 0.013 |
5.221 ± 0.036 | 7.814 ± 0.054 |
4.53 ± 0.037 | 6.945 ± 0.053 |
2.169 ± 0.02 | 7.477 ± 0.046 |
6.392 ± 0.039 | 9.81 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.85 ± 0.022 | 4.082 ± 0.028 |
3.657 ± 0.024 | 3.835 ± 0.029 |
4.118 ± 0.028 | 6.432 ± 0.033 |
5.45 ± 0.055 | 6.869 ± 0.035 |
1.087 ± 0.017 | 3.309 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
