
Tropicibacter phthalicicus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Tropicibacter
Average proteome isoelectric point is 6.01
Get precalculated fractions of proteins
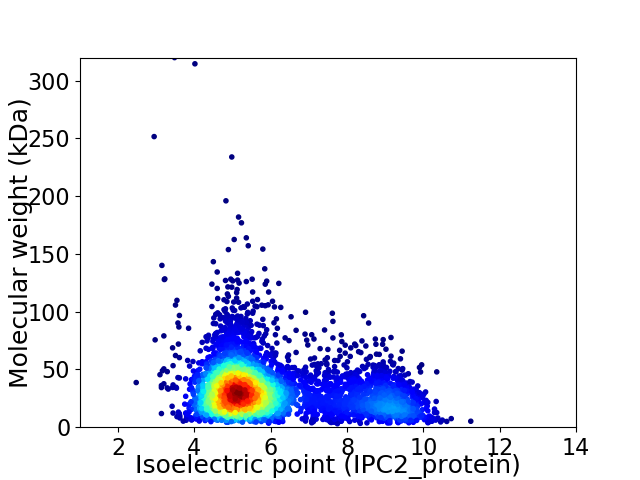
Virtual 2D-PAGE plot for 4708 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A238JC85|A0A238JC85_9RHOB RHH_4 domain-containing protein OS=Tropicibacter phthalicicus OX=1037362 GN=TRP8649_02426 PE=4 SV=1
MM1 pKa = 7.33PTINHH6 pKa = 7.13DD7 pKa = 3.57PTALTTSQGDD17 pKa = 3.44VTSAHH22 pKa = 6.47FGSNFLFDD30 pKa = 4.05QDD32 pKa = 4.39FNPDD36 pKa = 2.97GTIEE40 pKa = 4.28GNWYY44 pKa = 9.89DD45 pKa = 3.79QVTDD49 pKa = 3.68AAHH52 pKa = 5.58TQTLRR57 pKa = 11.84FPGGTITEE65 pKa = 4.42TQFDD69 pKa = 4.67LNDD72 pKa = 3.62TAKK75 pKa = 10.92LEE77 pKa = 4.23EE78 pKa = 4.09FLSYY82 pKa = 10.81CSANGHH88 pKa = 6.36KK89 pKa = 10.72AMIVLPTFRR98 pKa = 11.84FFDD101 pKa = 4.47PDD103 pKa = 3.84SPNSNYY109 pKa = 10.33LEE111 pKa = 4.38SDD113 pKa = 3.22AEE115 pKa = 4.33TIIKK119 pKa = 10.58NFVQTVLDD127 pKa = 4.46LADD130 pKa = 3.78AQNPKK135 pKa = 10.48VIIEE139 pKa = 4.02GFEE142 pKa = 3.8IGNEE146 pKa = 4.06YY147 pKa = 8.95YY148 pKa = 10.13QKK150 pKa = 11.26KK151 pKa = 7.7FTNEE155 pKa = 3.54DD156 pKa = 3.41DD157 pKa = 4.81LLFGDD162 pKa = 4.27EE163 pKa = 4.98DD164 pKa = 4.15NPFVADD170 pKa = 5.08EE171 pKa = 4.1DD172 pKa = 4.85TPWTTEE178 pKa = 3.46QFARR182 pKa = 11.84LQAQMASWVNDD193 pKa = 3.23VVADD197 pKa = 3.79NVGVYY202 pKa = 8.69VQAGRR207 pKa = 11.84NSDD210 pKa = 3.93EE211 pKa = 4.13NDD213 pKa = 3.17TLYY216 pKa = 11.35GGFADD221 pKa = 5.08AGIGLDD227 pKa = 3.9VIDD230 pKa = 5.06GVLTHH235 pKa = 7.12LYY237 pKa = 9.84YY238 pKa = 10.32PPEE241 pKa = 4.14NRR243 pKa = 11.84DD244 pKa = 3.1LSVIEE249 pKa = 4.13DD250 pKa = 3.59VEE252 pKa = 4.34VRR254 pKa = 11.84IEE256 pKa = 4.14TIRR259 pKa = 11.84EE260 pKa = 3.68VWGSSRR266 pKa = 11.84EE267 pKa = 4.15LIVTEE272 pKa = 4.45WNTSEE277 pKa = 3.84ANNYY281 pKa = 8.85GGNNAVYY288 pKa = 10.69GMARR292 pKa = 11.84LAALTRR298 pKa = 11.84GFADD302 pKa = 5.17FIGHH306 pKa = 6.75GVDD309 pKa = 5.17QLLIWSTIAGGPNGRR324 pKa = 11.84ATLGLHH330 pKa = 5.04TTAEE334 pKa = 4.33QVFTPTGYY342 pKa = 9.79WFRR345 pKa = 11.84LMASEE350 pKa = 6.22LIDD353 pKa = 3.56TQLVSDD359 pKa = 4.31YY360 pKa = 11.16GSASRR365 pKa = 11.84LLDD368 pKa = 3.81AQDD371 pKa = 3.52EE372 pKa = 4.77FVGYY376 pKa = 10.36SYY378 pKa = 11.46VFQGTTFEE386 pKa = 4.38TSNDD390 pKa = 3.32YY391 pKa = 11.01HH392 pKa = 8.41LLYY395 pKa = 11.26SNGTNGDD402 pKa = 3.55LTLDD406 pKa = 3.96LDD408 pKa = 5.09LSNLMTNDD416 pKa = 2.98SFVYY420 pKa = 9.87AAHH423 pKa = 6.67IVHH426 pKa = 6.89TGALSQGDD434 pKa = 4.18PQDD437 pKa = 3.87AADD440 pKa = 4.47PADD443 pKa = 3.56PHH445 pKa = 6.13ATAEE449 pKa = 4.26LQFVTNFEE457 pKa = 4.27DD458 pKa = 5.38LEE460 pKa = 5.56DD461 pKa = 4.4YY462 pKa = 11.39DD463 pKa = 4.08LGAYY467 pKa = 8.29EE468 pKa = 4.4FLQVNIVTGHH478 pKa = 6.61GIDD481 pKa = 5.44LEE483 pKa = 4.29TDD485 pKa = 3.5PFNAVDD491 pKa = 5.01DD492 pKa = 4.34NMSLTKK498 pKa = 10.9YY499 pKa = 10.49GDD501 pKa = 3.51RR502 pKa = 11.84LSTFDD507 pKa = 4.94GNDD510 pKa = 3.09SVYY513 pKa = 11.06GGAGFDD519 pKa = 3.86TVDD522 pKa = 4.03GGDD525 pKa = 3.66GNDD528 pKa = 4.47FINGEE533 pKa = 4.03NHH535 pKa = 7.03ADD537 pKa = 3.68SLLGGAGDD545 pKa = 3.74DD546 pKa = 3.98TLIGGHH552 pKa = 6.4GVDD555 pKa = 3.7QLFGGVGHH563 pKa = 7.67DD564 pKa = 3.77SLDD567 pKa = 3.79AGSEE571 pKa = 3.91ADD573 pKa = 4.44RR574 pKa = 11.84VWGGDD579 pKa = 3.15GNDD582 pKa = 3.84IIRR585 pKa = 11.84AGSNFGEE592 pKa = 4.65SVDD595 pKa = 4.08GLWGEE600 pKa = 4.5AGNDD604 pKa = 3.57TLYY607 pKa = 11.5GDD609 pKa = 4.56AGFDD613 pKa = 3.7YY614 pKa = 11.11LSGGTGEE621 pKa = 4.78DD622 pKa = 3.62VLIGGDD628 pKa = 3.63QADD631 pKa = 4.3NLYY634 pKa = 11.02GDD636 pKa = 4.82AGNDD640 pKa = 3.32SLYY643 pKa = 11.02GGQGLDD649 pKa = 3.59RR650 pKa = 11.84MFGGDD655 pKa = 3.78GNDD658 pKa = 3.43YY659 pKa = 10.75LYY661 pKa = 10.68DD662 pKa = 3.49ITDD665 pKa = 3.42SNGFFGEE672 pKa = 4.58GGDD675 pKa = 4.19DD676 pKa = 3.64MMVAGNGNDD685 pKa = 3.08RR686 pKa = 11.84FFGGTGNDD694 pKa = 3.46IISAGGGNDD703 pKa = 3.67SIYY706 pKa = 11.07GGAGFDD712 pKa = 4.14TIIGGAGNDD721 pKa = 3.58VMRR724 pKa = 11.84GDD726 pKa = 4.49FNADD730 pKa = 3.06TFVFEE735 pKa = 5.61DD736 pKa = 3.52GHH738 pKa = 7.89GSDD741 pKa = 4.79TISDD745 pKa = 3.94FDD747 pKa = 4.33TNTNLEE753 pKa = 4.16QIDD756 pKa = 4.22LSGVSAIANVSDD768 pKa = 4.69LNLGSATSGAATQVGADD785 pKa = 3.45VVINTGGGNSIRR797 pKa = 11.84LTGVNIADD805 pKa = 4.41LEE807 pKa = 4.59AGDD810 pKa = 5.36FIFF813 pKa = 5.87
MM1 pKa = 7.33PTINHH6 pKa = 7.13DD7 pKa = 3.57PTALTTSQGDD17 pKa = 3.44VTSAHH22 pKa = 6.47FGSNFLFDD30 pKa = 4.05QDD32 pKa = 4.39FNPDD36 pKa = 2.97GTIEE40 pKa = 4.28GNWYY44 pKa = 9.89DD45 pKa = 3.79QVTDD49 pKa = 3.68AAHH52 pKa = 5.58TQTLRR57 pKa = 11.84FPGGTITEE65 pKa = 4.42TQFDD69 pKa = 4.67LNDD72 pKa = 3.62TAKK75 pKa = 10.92LEE77 pKa = 4.23EE78 pKa = 4.09FLSYY82 pKa = 10.81CSANGHH88 pKa = 6.36KK89 pKa = 10.72AMIVLPTFRR98 pKa = 11.84FFDD101 pKa = 4.47PDD103 pKa = 3.84SPNSNYY109 pKa = 10.33LEE111 pKa = 4.38SDD113 pKa = 3.22AEE115 pKa = 4.33TIIKK119 pKa = 10.58NFVQTVLDD127 pKa = 4.46LADD130 pKa = 3.78AQNPKK135 pKa = 10.48VIIEE139 pKa = 4.02GFEE142 pKa = 3.8IGNEE146 pKa = 4.06YY147 pKa = 8.95YY148 pKa = 10.13QKK150 pKa = 11.26KK151 pKa = 7.7FTNEE155 pKa = 3.54DD156 pKa = 3.41DD157 pKa = 4.81LLFGDD162 pKa = 4.27EE163 pKa = 4.98DD164 pKa = 4.15NPFVADD170 pKa = 5.08EE171 pKa = 4.1DD172 pKa = 4.85TPWTTEE178 pKa = 3.46QFARR182 pKa = 11.84LQAQMASWVNDD193 pKa = 3.23VVADD197 pKa = 3.79NVGVYY202 pKa = 8.69VQAGRR207 pKa = 11.84NSDD210 pKa = 3.93EE211 pKa = 4.13NDD213 pKa = 3.17TLYY216 pKa = 11.35GGFADD221 pKa = 5.08AGIGLDD227 pKa = 3.9VIDD230 pKa = 5.06GVLTHH235 pKa = 7.12LYY237 pKa = 9.84YY238 pKa = 10.32PPEE241 pKa = 4.14NRR243 pKa = 11.84DD244 pKa = 3.1LSVIEE249 pKa = 4.13DD250 pKa = 3.59VEE252 pKa = 4.34VRR254 pKa = 11.84IEE256 pKa = 4.14TIRR259 pKa = 11.84EE260 pKa = 3.68VWGSSRR266 pKa = 11.84EE267 pKa = 4.15LIVTEE272 pKa = 4.45WNTSEE277 pKa = 3.84ANNYY281 pKa = 8.85GGNNAVYY288 pKa = 10.69GMARR292 pKa = 11.84LAALTRR298 pKa = 11.84GFADD302 pKa = 5.17FIGHH306 pKa = 6.75GVDD309 pKa = 5.17QLLIWSTIAGGPNGRR324 pKa = 11.84ATLGLHH330 pKa = 5.04TTAEE334 pKa = 4.33QVFTPTGYY342 pKa = 9.79WFRR345 pKa = 11.84LMASEE350 pKa = 6.22LIDD353 pKa = 3.56TQLVSDD359 pKa = 4.31YY360 pKa = 11.16GSASRR365 pKa = 11.84LLDD368 pKa = 3.81AQDD371 pKa = 3.52EE372 pKa = 4.77FVGYY376 pKa = 10.36SYY378 pKa = 11.46VFQGTTFEE386 pKa = 4.38TSNDD390 pKa = 3.32YY391 pKa = 11.01HH392 pKa = 8.41LLYY395 pKa = 11.26SNGTNGDD402 pKa = 3.55LTLDD406 pKa = 3.96LDD408 pKa = 5.09LSNLMTNDD416 pKa = 2.98SFVYY420 pKa = 9.87AAHH423 pKa = 6.67IVHH426 pKa = 6.89TGALSQGDD434 pKa = 4.18PQDD437 pKa = 3.87AADD440 pKa = 4.47PADD443 pKa = 3.56PHH445 pKa = 6.13ATAEE449 pKa = 4.26LQFVTNFEE457 pKa = 4.27DD458 pKa = 5.38LEE460 pKa = 5.56DD461 pKa = 4.4YY462 pKa = 11.39DD463 pKa = 4.08LGAYY467 pKa = 8.29EE468 pKa = 4.4FLQVNIVTGHH478 pKa = 6.61GIDD481 pKa = 5.44LEE483 pKa = 4.29TDD485 pKa = 3.5PFNAVDD491 pKa = 5.01DD492 pKa = 4.34NMSLTKK498 pKa = 10.9YY499 pKa = 10.49GDD501 pKa = 3.51RR502 pKa = 11.84LSTFDD507 pKa = 4.94GNDD510 pKa = 3.09SVYY513 pKa = 11.06GGAGFDD519 pKa = 3.86TVDD522 pKa = 4.03GGDD525 pKa = 3.66GNDD528 pKa = 4.47FINGEE533 pKa = 4.03NHH535 pKa = 7.03ADD537 pKa = 3.68SLLGGAGDD545 pKa = 3.74DD546 pKa = 3.98TLIGGHH552 pKa = 6.4GVDD555 pKa = 3.7QLFGGVGHH563 pKa = 7.67DD564 pKa = 3.77SLDD567 pKa = 3.79AGSEE571 pKa = 3.91ADD573 pKa = 4.44RR574 pKa = 11.84VWGGDD579 pKa = 3.15GNDD582 pKa = 3.84IIRR585 pKa = 11.84AGSNFGEE592 pKa = 4.65SVDD595 pKa = 4.08GLWGEE600 pKa = 4.5AGNDD604 pKa = 3.57TLYY607 pKa = 11.5GDD609 pKa = 4.56AGFDD613 pKa = 3.7YY614 pKa = 11.11LSGGTGEE621 pKa = 4.78DD622 pKa = 3.62VLIGGDD628 pKa = 3.63QADD631 pKa = 4.3NLYY634 pKa = 11.02GDD636 pKa = 4.82AGNDD640 pKa = 3.32SLYY643 pKa = 11.02GGQGLDD649 pKa = 3.59RR650 pKa = 11.84MFGGDD655 pKa = 3.78GNDD658 pKa = 3.43YY659 pKa = 10.75LYY661 pKa = 10.68DD662 pKa = 3.49ITDD665 pKa = 3.42SNGFFGEE672 pKa = 4.58GGDD675 pKa = 4.19DD676 pKa = 3.64MMVAGNGNDD685 pKa = 3.08RR686 pKa = 11.84FFGGTGNDD694 pKa = 3.46IISAGGGNDD703 pKa = 3.67SIYY706 pKa = 11.07GGAGFDD712 pKa = 4.14TIIGGAGNDD721 pKa = 3.58VMRR724 pKa = 11.84GDD726 pKa = 4.49FNADD730 pKa = 3.06TFVFEE735 pKa = 5.61DD736 pKa = 3.52GHH738 pKa = 7.89GSDD741 pKa = 4.79TISDD745 pKa = 3.94FDD747 pKa = 4.33TNTNLEE753 pKa = 4.16QIDD756 pKa = 4.22LSGVSAIANVSDD768 pKa = 4.69LNLGSATSGAATQVGADD785 pKa = 3.45VVINTGGGNSIRR797 pKa = 11.84LTGVNIADD805 pKa = 4.41LEE807 pKa = 4.59AGDD810 pKa = 5.36FIFF813 pKa = 5.87
Molecular weight: 86.76 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A238JIC0|A0A238JIC0_9RHOB Rieske [2Fe-2S] domain protein OS=Tropicibacter phthalicicus OX=1037362 GN=TRP8649_04037 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.62GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.45AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.62GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.45AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1437188 |
29 |
3208 |
305.3 |
33.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.541 ± 0.045 | 0.952 ± 0.013 |
6.146 ± 0.039 | 6.121 ± 0.033 |
3.877 ± 0.021 | 8.462 ± 0.044 |
2.093 ± 0.02 | 5.221 ± 0.027 |
3.655 ± 0.028 | 10.038 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.801 ± 0.018 | 2.858 ± 0.019 |
4.894 ± 0.027 | 3.589 ± 0.02 |
6.053 ± 0.035 | 5.525 ± 0.029 |
5.404 ± 0.033 | 7.104 ± 0.03 |
1.412 ± 0.015 | 2.254 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
