
Thogoto virus (isolate SiAr 126) (Tho)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Insthoviricetes; Articulavirales; Orthomyxoviridae; Thogotovirus; Thogoto thogotovirus
Average proteome isoelectric point is 6.38
Get precalculated fractions of proteins
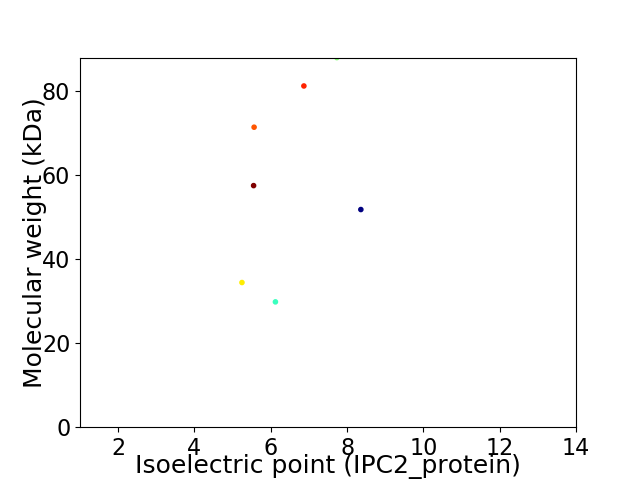
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q9YNA4|PB2_THOGV Polymerase basic protein 2 OS=Thogoto virus (isolate SiAr 126) OX=126796 GN=Segment 1 PE=1 SV=1
MM1 pKa = 7.76ASNLPVRR8 pKa = 11.84SFSEE12 pKa = 4.51VCCAEE17 pKa = 3.5ARR19 pKa = 11.84AAIIQMEE26 pKa = 4.47NNPDD30 pKa = 3.21EE31 pKa = 4.62TVCNRR36 pKa = 11.84IWKK39 pKa = 8.82IHH41 pKa = 7.02RR42 pKa = 11.84DD43 pKa = 3.71LQSSDD48 pKa = 3.19LTTTVQVMMVYY59 pKa = 10.28RR60 pKa = 11.84FISKK64 pKa = 10.02RR65 pKa = 11.84VPEE68 pKa = 4.45GCFAILSGVNTGMYY82 pKa = 10.17NPRR85 pKa = 11.84EE86 pKa = 4.2LKK88 pKa = 10.28RR89 pKa = 11.84SYY91 pKa = 10.46VQSLSSGTSCEE102 pKa = 4.13FLRR105 pKa = 11.84SLDD108 pKa = 3.73KK109 pKa = 10.9LAKK112 pKa = 9.96NLLAVHH118 pKa = 6.15VCSDD122 pKa = 3.35VKK124 pKa = 10.67MSLNKK129 pKa = 10.09RR130 pKa = 11.84QVIDD134 pKa = 4.68FISGEE139 pKa = 4.18EE140 pKa = 4.31DD141 pKa = 3.34PTLHH145 pKa = 5.9TAEE148 pKa = 5.08HH149 pKa = 6.01LTSLALDD156 pKa = 4.42DD157 pKa = 4.56SPSAVVYY164 pKa = 10.53SGWQQEE170 pKa = 4.25AIKK173 pKa = 10.42LHH175 pKa = 4.32NTIRR179 pKa = 11.84KK180 pKa = 7.43IATMRR185 pKa = 11.84PADD188 pKa = 3.75CKK190 pKa = 10.8AGKK193 pKa = 9.65FYY195 pKa = 11.29SDD197 pKa = 3.78ILSACDD203 pKa = 3.26QTKK206 pKa = 10.49EE207 pKa = 4.09LLDD210 pKa = 4.42AFDD213 pKa = 3.87QGKK216 pKa = 8.86LAYY219 pKa = 10.17DD220 pKa = 3.57RR221 pKa = 11.84DD222 pKa = 3.94VVLIGWMDD230 pKa = 4.31EE231 pKa = 4.01IIKK234 pKa = 10.3IFSKK238 pKa = 10.52PDD240 pKa = 3.14YY241 pKa = 10.71LEE243 pKa = 4.46AKK245 pKa = 8.94GVSYY249 pKa = 10.55QVLKK253 pKa = 11.03NVSNKK258 pKa = 9.26VALLRR263 pKa = 11.84EE264 pKa = 4.53SIWWVTEE271 pKa = 3.56LDD273 pKa = 3.28GRR275 pKa = 11.84EE276 pKa = 3.77YY277 pKa = 11.39LFFDD281 pKa = 4.02EE282 pKa = 4.35SWYY285 pKa = 10.1LHH287 pKa = 5.71GMSAFSDD294 pKa = 4.06GVPGYY299 pKa = 10.91EE300 pKa = 4.2DD301 pKa = 4.82FIYY304 pKa = 10.86
MM1 pKa = 7.76ASNLPVRR8 pKa = 11.84SFSEE12 pKa = 4.51VCCAEE17 pKa = 3.5ARR19 pKa = 11.84AAIIQMEE26 pKa = 4.47NNPDD30 pKa = 3.21EE31 pKa = 4.62TVCNRR36 pKa = 11.84IWKK39 pKa = 8.82IHH41 pKa = 7.02RR42 pKa = 11.84DD43 pKa = 3.71LQSSDD48 pKa = 3.19LTTTVQVMMVYY59 pKa = 10.28RR60 pKa = 11.84FISKK64 pKa = 10.02RR65 pKa = 11.84VPEE68 pKa = 4.45GCFAILSGVNTGMYY82 pKa = 10.17NPRR85 pKa = 11.84EE86 pKa = 4.2LKK88 pKa = 10.28RR89 pKa = 11.84SYY91 pKa = 10.46VQSLSSGTSCEE102 pKa = 4.13FLRR105 pKa = 11.84SLDD108 pKa = 3.73KK109 pKa = 10.9LAKK112 pKa = 9.96NLLAVHH118 pKa = 6.15VCSDD122 pKa = 3.35VKK124 pKa = 10.67MSLNKK129 pKa = 10.09RR130 pKa = 11.84QVIDD134 pKa = 4.68FISGEE139 pKa = 4.18EE140 pKa = 4.31DD141 pKa = 3.34PTLHH145 pKa = 5.9TAEE148 pKa = 5.08HH149 pKa = 6.01LTSLALDD156 pKa = 4.42DD157 pKa = 4.56SPSAVVYY164 pKa = 10.53SGWQQEE170 pKa = 4.25AIKK173 pKa = 10.42LHH175 pKa = 4.32NTIRR179 pKa = 11.84KK180 pKa = 7.43IATMRR185 pKa = 11.84PADD188 pKa = 3.75CKK190 pKa = 10.8AGKK193 pKa = 9.65FYY195 pKa = 11.29SDD197 pKa = 3.78ILSACDD203 pKa = 3.26QTKK206 pKa = 10.49EE207 pKa = 4.09LLDD210 pKa = 4.42AFDD213 pKa = 3.87QGKK216 pKa = 8.86LAYY219 pKa = 10.17DD220 pKa = 3.57RR221 pKa = 11.84DD222 pKa = 3.94VVLIGWMDD230 pKa = 4.31EE231 pKa = 4.01IIKK234 pKa = 10.3IFSKK238 pKa = 10.52PDD240 pKa = 3.14YY241 pKa = 10.71LEE243 pKa = 4.46AKK245 pKa = 8.94GVSYY249 pKa = 10.55QVLKK253 pKa = 11.03NVSNKK258 pKa = 9.26VALLRR263 pKa = 11.84EE264 pKa = 4.53SIWWVTEE271 pKa = 3.56LDD273 pKa = 3.28GRR275 pKa = 11.84EE276 pKa = 3.77YY277 pKa = 11.39LFFDD281 pKa = 4.02EE282 pKa = 4.35SWYY285 pKa = 10.1LHH287 pKa = 5.71GMSAFSDD294 pKa = 4.06GVPGYY299 pKa = 10.91EE300 pKa = 4.2DD301 pKa = 4.82FIYY304 pKa = 10.86
Molecular weight: 34.46 kDa
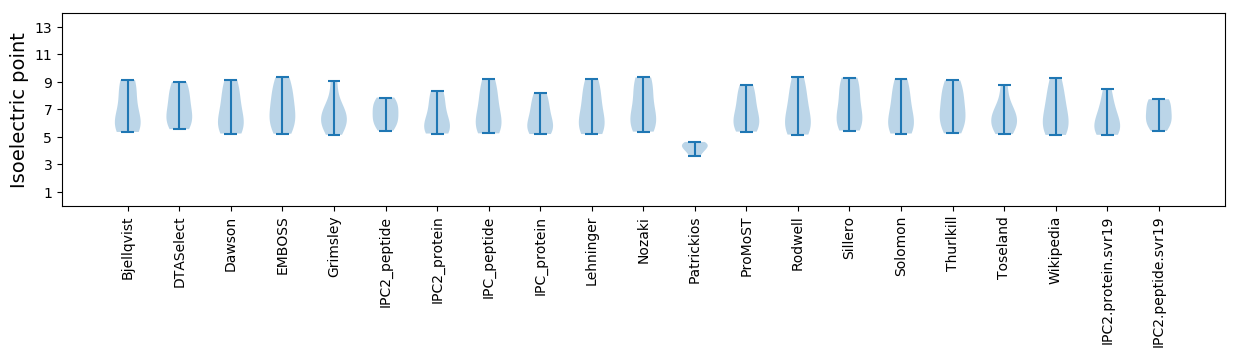
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q77D96|M1_THOGV Matrix protein OS=Thogoto virus (isolate SiAr 126) OX=126796 GN=Segment 6 PE=4 SV=1
MM1 pKa = 7.45ATDD4 pKa = 3.57QMDD7 pKa = 3.23ISGPPPKK14 pKa = 9.55KK15 pKa = 9.6QHH17 pKa = 6.12VDD19 pKa = 3.28TEE21 pKa = 4.41SQIPKK26 pKa = 8.4MYY28 pKa = 10.93EE29 pKa = 3.7MIRR32 pKa = 11.84DD33 pKa = 3.62QMRR36 pKa = 11.84TLASTHH42 pKa = 6.68KK43 pKa = 10.06IPLNIDD49 pKa = 3.42HH50 pKa = 6.66NCEE53 pKa = 4.18VIGSIIMAACTNNRR67 pKa = 11.84DD68 pKa = 3.52LRR70 pKa = 11.84PVDD73 pKa = 4.09KK74 pKa = 10.73YY75 pKa = 10.27WFLMGPAGAEE85 pKa = 3.79VMTEE89 pKa = 3.97VEE91 pKa = 5.28IDD93 pKa = 3.61IQPQLQWAKK102 pKa = 10.8GAVHH106 pKa = 6.7DD107 pKa = 4.83PKK109 pKa = 11.4YY110 pKa = 10.38KK111 pKa = 9.24GQWYY115 pKa = 8.51PFLALLQISNKK126 pKa = 9.26TKK128 pKa = 10.05DD129 pKa = 3.15TILWQKK135 pKa = 10.74YY136 pKa = 8.83PVTQEE141 pKa = 4.54LEE143 pKa = 3.83ISNSLEE149 pKa = 3.7IYY151 pKa = 11.01ANGHH155 pKa = 5.62GIKK158 pKa = 10.29DD159 pKa = 3.4RR160 pKa = 11.84LKK162 pKa = 9.09NSRR165 pKa = 11.84PRR167 pKa = 11.84SVGPLVHH174 pKa = 6.94LLHH177 pKa = 7.33LKK179 pKa = 10.17RR180 pKa = 11.84LQEE183 pKa = 4.25NPPKK187 pKa = 10.56NPKK190 pKa = 8.08TKK192 pKa = 10.5KK193 pKa = 9.27PLEE196 pKa = 4.39SPAVNGIRR204 pKa = 11.84KK205 pKa = 9.87SIVGHH210 pKa = 6.69LKK212 pKa = 9.68RR213 pKa = 11.84QCIGEE218 pKa = 4.24TQKK221 pKa = 11.76AMINQFEE228 pKa = 4.26MGRR231 pKa = 11.84WEE233 pKa = 4.22SLSTFAASLLAIKK246 pKa = 9.97PRR248 pKa = 11.84IEE250 pKa = 3.67NHH252 pKa = 5.91FVLTYY257 pKa = 9.88PLIANCEE264 pKa = 4.09DD265 pKa = 3.52FAGATLSDD273 pKa = 3.26EE274 pKa = 4.49WVFKK278 pKa = 11.14AMEE281 pKa = 4.86KK282 pKa = 10.17ISNKK286 pKa = 8.35KK287 pKa = 6.11TLRR290 pKa = 11.84VCGPDD295 pKa = 3.41EE296 pKa = 4.04KK297 pKa = 10.51WISFMNQIYY306 pKa = 9.56IHH308 pKa = 6.43SVFQTTGEE316 pKa = 4.02DD317 pKa = 3.25LGVLEE322 pKa = 4.73WVFGGRR328 pKa = 11.84FCQRR332 pKa = 11.84KK333 pKa = 8.8EE334 pKa = 3.73FGRR337 pKa = 11.84YY338 pKa = 8.32CKK340 pKa = 10.28KK341 pKa = 10.66SQTKK345 pKa = 10.48VIGLFTFQYY354 pKa = 9.89EE355 pKa = 4.54YY356 pKa = 10.22WSKK359 pKa = 10.31PLKK362 pKa = 9.97SAPRR366 pKa = 11.84SIEE369 pKa = 3.64GSKK372 pKa = 10.15RR373 pKa = 11.84GQISCRR379 pKa = 11.84PSFKK383 pKa = 10.57GKK385 pKa = 10.08RR386 pKa = 11.84PSYY389 pKa = 11.51NNFTSIDD396 pKa = 3.74ALQSASGSQTVSFYY410 pKa = 11.06DD411 pKa = 3.41QVRR414 pKa = 11.84EE415 pKa = 4.0EE416 pKa = 4.22CQKK419 pKa = 11.36YY420 pKa = 9.02MDD422 pKa = 4.74LKK424 pKa = 11.47VEE426 pKa = 4.2GTTCFYY432 pKa = 10.97RR433 pKa = 11.84KK434 pKa = 9.61GGHH437 pKa = 5.66VEE439 pKa = 4.01VEE441 pKa = 4.64FPGSAHH447 pKa = 6.57CNTYY451 pKa = 11.03LFGG454 pKa = 5.58
MM1 pKa = 7.45ATDD4 pKa = 3.57QMDD7 pKa = 3.23ISGPPPKK14 pKa = 9.55KK15 pKa = 9.6QHH17 pKa = 6.12VDD19 pKa = 3.28TEE21 pKa = 4.41SQIPKK26 pKa = 8.4MYY28 pKa = 10.93EE29 pKa = 3.7MIRR32 pKa = 11.84DD33 pKa = 3.62QMRR36 pKa = 11.84TLASTHH42 pKa = 6.68KK43 pKa = 10.06IPLNIDD49 pKa = 3.42HH50 pKa = 6.66NCEE53 pKa = 4.18VIGSIIMAACTNNRR67 pKa = 11.84DD68 pKa = 3.52LRR70 pKa = 11.84PVDD73 pKa = 4.09KK74 pKa = 10.73YY75 pKa = 10.27WFLMGPAGAEE85 pKa = 3.79VMTEE89 pKa = 3.97VEE91 pKa = 5.28IDD93 pKa = 3.61IQPQLQWAKK102 pKa = 10.8GAVHH106 pKa = 6.7DD107 pKa = 4.83PKK109 pKa = 11.4YY110 pKa = 10.38KK111 pKa = 9.24GQWYY115 pKa = 8.51PFLALLQISNKK126 pKa = 9.26TKK128 pKa = 10.05DD129 pKa = 3.15TILWQKK135 pKa = 10.74YY136 pKa = 8.83PVTQEE141 pKa = 4.54LEE143 pKa = 3.83ISNSLEE149 pKa = 3.7IYY151 pKa = 11.01ANGHH155 pKa = 5.62GIKK158 pKa = 10.29DD159 pKa = 3.4RR160 pKa = 11.84LKK162 pKa = 9.09NSRR165 pKa = 11.84PRR167 pKa = 11.84SVGPLVHH174 pKa = 6.94LLHH177 pKa = 7.33LKK179 pKa = 10.17RR180 pKa = 11.84LQEE183 pKa = 4.25NPPKK187 pKa = 10.56NPKK190 pKa = 8.08TKK192 pKa = 10.5KK193 pKa = 9.27PLEE196 pKa = 4.39SPAVNGIRR204 pKa = 11.84KK205 pKa = 9.87SIVGHH210 pKa = 6.69LKK212 pKa = 9.68RR213 pKa = 11.84QCIGEE218 pKa = 4.24TQKK221 pKa = 11.76AMINQFEE228 pKa = 4.26MGRR231 pKa = 11.84WEE233 pKa = 4.22SLSTFAASLLAIKK246 pKa = 9.97PRR248 pKa = 11.84IEE250 pKa = 3.67NHH252 pKa = 5.91FVLTYY257 pKa = 9.88PLIANCEE264 pKa = 4.09DD265 pKa = 3.52FAGATLSDD273 pKa = 3.26EE274 pKa = 4.49WVFKK278 pKa = 11.14AMEE281 pKa = 4.86KK282 pKa = 10.17ISNKK286 pKa = 8.35KK287 pKa = 6.11TLRR290 pKa = 11.84VCGPDD295 pKa = 3.41EE296 pKa = 4.04KK297 pKa = 10.51WISFMNQIYY306 pKa = 9.56IHH308 pKa = 6.43SVFQTTGEE316 pKa = 4.02DD317 pKa = 3.25LGVLEE322 pKa = 4.73WVFGGRR328 pKa = 11.84FCQRR332 pKa = 11.84KK333 pKa = 8.8EE334 pKa = 3.73FGRR337 pKa = 11.84YY338 pKa = 8.32CKK340 pKa = 10.28KK341 pKa = 10.66SQTKK345 pKa = 10.48VIGLFTFQYY354 pKa = 9.89EE355 pKa = 4.54YY356 pKa = 10.22WSKK359 pKa = 10.31PLKK362 pKa = 9.97SAPRR366 pKa = 11.84SIEE369 pKa = 3.64GSKK372 pKa = 10.15RR373 pKa = 11.84GQISCRR379 pKa = 11.84PSFKK383 pKa = 10.57GKK385 pKa = 10.08RR386 pKa = 11.84PSYY389 pKa = 11.51NNFTSIDD396 pKa = 3.74ALQSASGSQTVSFYY410 pKa = 11.06DD411 pKa = 3.41QVRR414 pKa = 11.84EE415 pKa = 4.0EE416 pKa = 4.22CQKK419 pKa = 11.36YY420 pKa = 9.02MDD422 pKa = 4.74LKK424 pKa = 11.47VEE426 pKa = 4.2GTTCFYY432 pKa = 10.97RR433 pKa = 11.84KK434 pKa = 9.61GGHH437 pKa = 5.66VEE439 pKa = 4.01VEE441 pKa = 4.64FPGSAHH447 pKa = 6.57CNTYY451 pKa = 11.03LFGG454 pKa = 5.58
Molecular weight: 51.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3637 |
266 |
769 |
519.6 |
59.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.801 ± 0.364 | 2.337 ± 0.236 |
5.334 ± 0.405 | 7.231 ± 0.552 |
3.932 ± 0.336 | 5.307 ± 0.367 |
2.392 ± 0.098 | 5.829 ± 0.35 |
6.571 ± 0.445 | 9.211 ± 0.486 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.64 ± 0.339 | 3.987 ± 0.269 |
4.839 ± 0.415 | 3.822 ± 0.368 |
5.856 ± 0.432 | 7.451 ± 0.519 |
5.884 ± 0.372 | 6.379 ± 0.371 |
1.705 ± 0.327 | 3.492 ± 0.207 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
