
Saccharolobus solfataricus (strain ATCC 35092 / DSM 1617 / JCM 11322 / P2) (Sulfolobus solfataricus)
Taxonomy: cellular organisms; Archaea; TACK group; Crenarchaeota; Thermoprotei; Sulfolobales; Sulfolobaceae; Saccharolobus; Saccharolobus solfataricus
Average proteome isoelectric point is 7.2
Get precalculated fractions of proteins
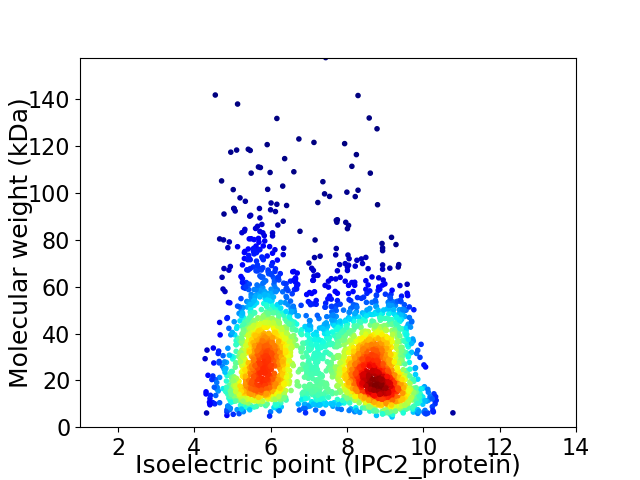
Virtual 2D-PAGE plot for 2938 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q97U05|Q97U05_SACS2 Amino acid transporter related protein OS=Saccharolobus solfataricus (strain ATCC 35092 / DSM 1617 / JCM 11322 / P2) OX=273057 GN=SSO3224 PE=4 SV=1
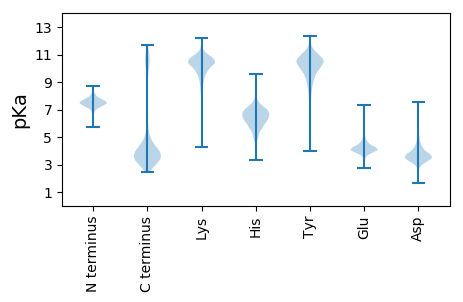
MM1 pKa = 7.03EE2 pKa = 5.07TLNEE6 pKa = 4.33SKK8 pKa = 10.83KK9 pKa = 10.5EE10 pKa = 3.86FYY12 pKa = 10.17TYY14 pKa = 10.64FISTSKK20 pKa = 10.56FYY22 pKa = 11.31YY23 pKa = 10.39DD24 pKa = 4.74LSNTVNSPIVVCEE37 pKa = 3.85MLYY40 pKa = 10.41EE41 pKa = 4.89AINAGIKK48 pKa = 9.97LLSYY52 pKa = 10.52YY53 pKa = 10.61FSLQDD58 pKa = 3.23KK59 pKa = 10.1PRR61 pKa = 11.84TEE63 pKa = 4.19VVKK66 pKa = 10.29EE67 pKa = 3.8LSSILGDD74 pKa = 3.11WVEE77 pKa = 4.5YY78 pKa = 9.71YY79 pKa = 10.07WNLGLTLHH87 pKa = 6.24YY88 pKa = 10.73DD89 pKa = 3.99CYY91 pKa = 11.34LSGNVDD97 pKa = 3.37EE98 pKa = 6.27DD99 pKa = 5.64DD100 pKa = 3.61IPLYY104 pKa = 10.45EE105 pKa = 4.19NQVKK109 pKa = 10.64DD110 pKa = 4.47FISRR114 pKa = 11.84VEE116 pKa = 3.96EE117 pKa = 4.25VVFGG121 pKa = 3.88
MM1 pKa = 7.03EE2 pKa = 5.07TLNEE6 pKa = 4.33SKK8 pKa = 10.83KK9 pKa = 10.5EE10 pKa = 3.86FYY12 pKa = 10.17TYY14 pKa = 10.64FISTSKK20 pKa = 10.56FYY22 pKa = 11.31YY23 pKa = 10.39DD24 pKa = 4.74LSNTVNSPIVVCEE37 pKa = 3.85MLYY40 pKa = 10.41EE41 pKa = 4.89AINAGIKK48 pKa = 9.97LLSYY52 pKa = 10.52YY53 pKa = 10.61FSLQDD58 pKa = 3.23KK59 pKa = 10.1PRR61 pKa = 11.84TEE63 pKa = 4.19VVKK66 pKa = 10.29EE67 pKa = 3.8LSSILGDD74 pKa = 3.11WVEE77 pKa = 4.5YY78 pKa = 9.71YY79 pKa = 10.07WNLGLTLHH87 pKa = 6.24YY88 pKa = 10.73DD89 pKa = 3.99CYY91 pKa = 11.34LSGNVDD97 pKa = 3.37EE98 pKa = 6.27DD99 pKa = 5.64DD100 pKa = 3.61IPLYY104 pKa = 10.45EE105 pKa = 4.19NQVKK109 pKa = 10.64DD110 pKa = 4.47FISRR114 pKa = 11.84VEE116 pKa = 3.96EE117 pKa = 4.25VVFGG121 pKa = 3.88
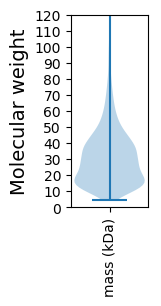
Molecular weight: 14.21 kDa
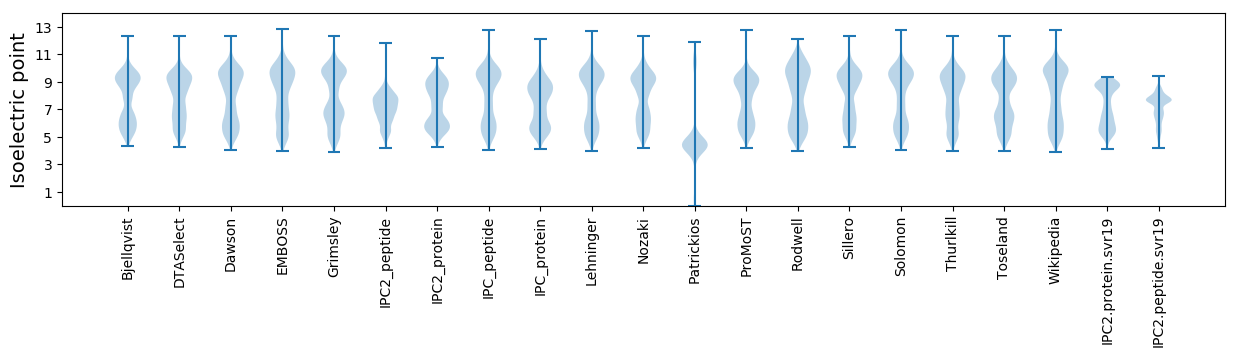
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q97YK4|Q97YK4_SACS2 Second ORF in transposon ISC1316 OS=Saccharolobus solfataricus (strain ATCC 35092 / DSM 1617 / JCM 11322 / P2) OX=273057 GN=SSO7675 PE=4 SV=1
MM1 pKa = 6.95HH2 pKa = 6.96QSTRR6 pKa = 11.84RR7 pKa = 11.84GRR9 pKa = 11.84EE10 pKa = 3.58LMRR13 pKa = 11.84KK14 pKa = 8.29YY15 pKa = 10.44SHH17 pKa = 7.0RR18 pKa = 11.84EE19 pKa = 3.37RR20 pKa = 11.84NRR22 pKa = 11.84VLDD25 pKa = 4.36FVHH28 pKa = 6.84KK29 pKa = 10.11FVNTLLDD36 pKa = 4.62LYY38 pKa = 10.18PMTFFAVEE46 pKa = 4.13KK47 pKa = 10.36LNKK50 pKa = 10.05EE51 pKa = 4.6SMFKK55 pKa = 10.61DD56 pKa = 3.43ANDD59 pKa = 3.51SLSRR63 pKa = 11.84KK64 pKa = 9.09ISRR67 pKa = 11.84TVWRR71 pKa = 11.84SIHH74 pKa = 6.78RR75 pKa = 11.84VLKK78 pKa = 10.63YY79 pKa = 9.94KK80 pKa = 10.85APLYY84 pKa = 10.4GSFVKK89 pKa = 10.31EE90 pKa = 3.71VNPYY94 pKa = 8.3LTSRR98 pKa = 11.84SCPRR102 pKa = 11.84CRR104 pKa = 11.84FVSRR108 pKa = 11.84KK109 pKa = 8.54VGKK112 pKa = 7.94TFEE115 pKa = 4.21CEE117 pKa = 3.44RR118 pKa = 11.84CGFKK122 pKa = 10.55LDD124 pKa = 3.66RR125 pKa = 11.84QLNASLNIYY134 pKa = 10.63LKK136 pKa = 9.76MCGFPHH142 pKa = 6.88IRR144 pKa = 11.84EE145 pKa = 3.97IARR148 pKa = 11.84VWVGVIPLMGRR159 pKa = 11.84RR160 pKa = 11.84RR161 pKa = 11.84MNVRR165 pKa = 11.84DD166 pKa = 3.95FGEE169 pKa = 4.36AQGLRR174 pKa = 11.84VDD176 pKa = 4.17IKK178 pKa = 10.44YY179 pKa = 10.59HH180 pKa = 6.42EE181 pKa = 4.48IPP183 pKa = 4.03
MM1 pKa = 6.95HH2 pKa = 6.96QSTRR6 pKa = 11.84RR7 pKa = 11.84GRR9 pKa = 11.84EE10 pKa = 3.58LMRR13 pKa = 11.84KK14 pKa = 8.29YY15 pKa = 10.44SHH17 pKa = 7.0RR18 pKa = 11.84EE19 pKa = 3.37RR20 pKa = 11.84NRR22 pKa = 11.84VLDD25 pKa = 4.36FVHH28 pKa = 6.84KK29 pKa = 10.11FVNTLLDD36 pKa = 4.62LYY38 pKa = 10.18PMTFFAVEE46 pKa = 4.13KK47 pKa = 10.36LNKK50 pKa = 10.05EE51 pKa = 4.6SMFKK55 pKa = 10.61DD56 pKa = 3.43ANDD59 pKa = 3.51SLSRR63 pKa = 11.84KK64 pKa = 9.09ISRR67 pKa = 11.84TVWRR71 pKa = 11.84SIHH74 pKa = 6.78RR75 pKa = 11.84VLKK78 pKa = 10.63YY79 pKa = 9.94KK80 pKa = 10.85APLYY84 pKa = 10.4GSFVKK89 pKa = 10.31EE90 pKa = 3.71VNPYY94 pKa = 8.3LTSRR98 pKa = 11.84SCPRR102 pKa = 11.84CRR104 pKa = 11.84FVSRR108 pKa = 11.84KK109 pKa = 8.54VGKK112 pKa = 7.94TFEE115 pKa = 4.21CEE117 pKa = 3.44RR118 pKa = 11.84CGFKK122 pKa = 10.55LDD124 pKa = 3.66RR125 pKa = 11.84QLNASLNIYY134 pKa = 10.63LKK136 pKa = 9.76MCGFPHH142 pKa = 6.88IRR144 pKa = 11.84EE145 pKa = 3.97IARR148 pKa = 11.84VWVGVIPLMGRR159 pKa = 11.84RR160 pKa = 11.84RR161 pKa = 11.84MNVRR165 pKa = 11.84DD166 pKa = 3.95FGEE169 pKa = 4.36AQGLRR174 pKa = 11.84VDD176 pKa = 4.17IKK178 pKa = 10.44YY179 pKa = 10.59HH180 pKa = 6.42EE181 pKa = 4.48IPP183 pKa = 4.03
Molecular weight: 21.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
827883 |
40 |
1426 |
281.8 |
31.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.621 ± 0.05 | 0.618 ± 0.015 |
4.694 ± 0.038 | 6.807 ± 0.054 |
4.434 ± 0.036 | 6.44 ± 0.038 |
1.294 ± 0.016 | 9.5 ± 0.054 |
7.692 ± 0.061 | 10.356 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.201 ± 0.018 | 4.977 ± 0.041 |
3.814 ± 0.032 | 2.102 ± 0.025 |
4.684 ± 0.043 | 6.716 ± 0.041 |
4.734 ± 0.039 | 7.448 ± 0.035 |
1.049 ± 0.018 | 4.82 ± 0.036 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
