
Acinetobacter stercoris
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Moraxellaceae; Acinetobacter
Average proteome isoelectric point is 6.51
Get precalculated fractions of proteins
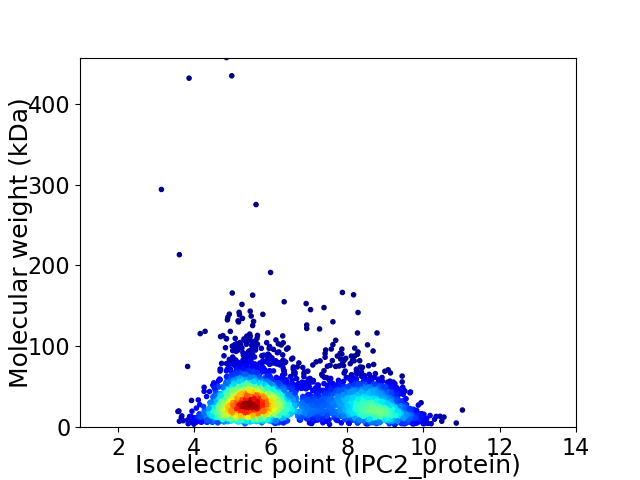
Virtual 2D-PAGE plot for 3857 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2U3N1N8|A0A2U3N1N8_9GAMM Phosphatidylglycerol--prolipoprotein diacylglyceryl transferase OS=Acinetobacter stercoris OX=2126983 GN=lgt PE=3 SV=1
MM1 pKa = 7.37GKK3 pKa = 8.14VTATDD8 pKa = 3.82GAGNTGPEE16 pKa = 4.25ATTGAADD23 pKa = 3.4VTAPVAPTVDD33 pKa = 3.55QNNAAGLGGTTEE45 pKa = 4.19PNAVITVEE53 pKa = 4.36LGNGSTVTTTADD65 pKa = 3.42SQGNWLLEE73 pKa = 4.28PNPLAEE79 pKa = 4.35GEE81 pKa = 4.43VGKK84 pKa = 8.63VTATDD89 pKa = 3.8GAGNSSTPTMTDD101 pKa = 2.73GTDD104 pKa = 2.75ITAPVAPTVDD114 pKa = 3.55HH115 pKa = 6.82NNAAGLGGTTEE126 pKa = 4.19PNAVITVEE134 pKa = 4.36LGNGSTVTTTADD146 pKa = 3.48SQGNWNIKK154 pKa = 9.15PNPIAEE160 pKa = 4.5GEE162 pKa = 4.4SATVIATDD170 pKa = 3.52KK171 pKa = 11.23AGNTSAEE178 pKa = 4.36TPTGKK183 pKa = 10.45ADD185 pKa = 3.77TVAPTAPGIDD195 pKa = 3.55QNNEE199 pKa = 3.73SGLSGTAEE207 pKa = 4.14PEE209 pKa = 4.03STISVEE215 pKa = 4.14LADD218 pKa = 4.02GTTITTTTDD227 pKa = 2.92TAGNWSITPNPLGDD241 pKa = 3.81GEE243 pKa = 4.53VGKK246 pKa = 8.62VTATDD251 pKa = 3.38AAGNNSAPTMTDD263 pKa = 2.7GTDD266 pKa = 3.19ITPPAAPGVDD276 pKa = 3.42QNNGEE281 pKa = 4.36GLSGTAEE288 pKa = 3.99AGATVTVKK296 pKa = 10.73DD297 pKa = 3.6ADD299 pKa = 4.11GNTVTTTADD308 pKa = 3.54STGHH312 pKa = 5.99WSITPNPISDD322 pKa = 3.67GMSGTVTATDD332 pKa = 3.45EE333 pKa = 4.79AGNEE337 pKa = 4.35SNPSNTGTADD347 pKa = 3.29TVAPNAPLVEE357 pKa = 4.24QNNAAGLAGTTEE369 pKa = 4.29PNAVITVDD377 pKa = 4.15LGNGSTVTTMADD389 pKa = 3.57SQGHH393 pKa = 5.15WSIEE397 pKa = 3.99PNPLEE402 pKa = 4.11TGEE405 pKa = 4.25VGKK408 pKa = 8.62VTATDD413 pKa = 3.42AAGNTSTPTMTDD425 pKa = 2.87GTDD428 pKa = 3.19ITPPAAPGIDD438 pKa = 3.42QNNGEE443 pKa = 4.58GLSGTAEE450 pKa = 3.99AGATVTVKK458 pKa = 10.73DD459 pKa = 3.6ADD461 pKa = 4.11GNTVTTTADD470 pKa = 3.54STGHH474 pKa = 5.99WSITPNPIADD484 pKa = 3.95GTSGTVTATDD494 pKa = 3.41EE495 pKa = 4.64AGNEE499 pKa = 4.12SSPSNTGTADD509 pKa = 3.29TVAPNAPLVEE519 pKa = 4.24QNNAAGLAGTTEE531 pKa = 4.29PNAVITVDD539 pKa = 4.15LGNGSTVTTMADD551 pKa = 3.57SQGHH555 pKa = 5.15WSIEE559 pKa = 3.99PNPLEE564 pKa = 4.11TGEE567 pKa = 4.25VGKK570 pKa = 8.62VTATDD575 pKa = 3.42AAGNTSTPTMTDD587 pKa = 2.87GTDD590 pKa = 3.19ITPPAAPGVDD600 pKa = 3.42QNNGEE605 pKa = 4.36GLSGTAEE612 pKa = 3.99AGATVTVKK620 pKa = 10.73DD621 pKa = 3.6ADD623 pKa = 4.11GNTVTTTADD632 pKa = 3.54STGHH636 pKa = 5.99WSITPNPITDD646 pKa = 3.61GSEE649 pKa = 4.22GSVTATDD656 pKa = 3.58AAGNTSAEE664 pKa = 4.55TPTGTADD671 pKa = 3.51TVAPTAPGIDD681 pKa = 3.53QNNGEE686 pKa = 4.58GLSGTAEE693 pKa = 3.99AGATVTVKK701 pKa = 10.73DD702 pKa = 3.6ADD704 pKa = 4.11GNTVTTTADD713 pKa = 3.54STGHH717 pKa = 5.99WSITPNPITDD727 pKa = 3.86GTSGTVTATDD737 pKa = 3.41EE738 pKa = 4.64AGNEE742 pKa = 4.12SSPSNTGIADD752 pKa = 4.02TVAPTAPGIDD762 pKa = 3.53QNNGEE767 pKa = 4.41GLSGTVEE774 pKa = 3.88AGATVIVKK782 pKa = 10.21DD783 pKa = 3.6ADD785 pKa = 3.83GNTVTTTADD794 pKa = 3.54STGHH798 pKa = 5.99WSITPNPLADD808 pKa = 3.81GSEE811 pKa = 4.35GSVTATDD818 pKa = 3.58AAGNTSAEE826 pKa = 4.55TPTGTADD833 pKa = 3.51TVAPTAPGIDD843 pKa = 3.53QNNGEE848 pKa = 4.58GLSGTAEE855 pKa = 3.99AGATVTVKK863 pKa = 10.73DD864 pKa = 3.6ADD866 pKa = 4.11GNTVTTTADD875 pKa = 3.54STGHH879 pKa = 5.99WSITPNPITDD889 pKa = 3.86GTSGTVTATDD899 pKa = 3.41EE900 pKa = 4.64AGNEE904 pKa = 4.05SSPTNTGIADD914 pKa = 4.18TVAPTAPGIDD924 pKa = 3.53QNNGEE929 pKa = 4.63GLTGTAEE936 pKa = 4.15AGSTVIVKK944 pKa = 10.26DD945 pKa = 3.34ADD947 pKa = 3.83GNTVTTTADD956 pKa = 3.54STGHH960 pKa = 5.99WSITPNPITDD970 pKa = 3.61GSEE973 pKa = 4.22GSVTATDD980 pKa = 3.58AAGNTSAEE988 pKa = 4.55TPTGTADD995 pKa = 4.48LVAPDD1000 pKa = 4.27KK1001 pKa = 11.15PLVNLVNGIDD1011 pKa = 4.65PITGTAEE1018 pKa = 3.88PNSIVTVTYY1027 pKa = 11.07LDD1029 pKa = 3.91GTSVTVTASSDD1040 pKa = 3.76GSWSVLNPGLEE1051 pKa = 4.58DD1052 pKa = 4.31GDD1054 pKa = 4.71TITVTATDD1062 pKa = 3.59EE1063 pKa = 4.24VGNKK1067 pKa = 9.61SEE1069 pKa = 4.43SVTVTVDD1076 pKa = 3.12TYY1078 pKa = 11.72VLADD1082 pKa = 3.45TVIITNGVTNDD1093 pKa = 3.57DD1094 pKa = 3.61TPAIVGTAEE1103 pKa = 3.99ANSIVTLTIIGTTGSQVVSVSVDD1126 pKa = 3.18GTGVWTYY1133 pKa = 10.24TPTVALAEE1141 pKa = 4.09DD1142 pKa = 5.12VYY1144 pKa = 10.53TLSAIARR1151 pKa = 11.84DD1152 pKa = 3.56SEE1154 pKa = 4.26GRR1156 pKa = 11.84VSQIGASSITVDD1168 pKa = 3.29TTPPSSEE1175 pKa = 3.47IVIEE1179 pKa = 4.02ITAITDD1185 pKa = 3.39DD1186 pKa = 4.21SGVLGDD1192 pKa = 5.9FITNDD1197 pKa = 3.61TTPTIHH1203 pKa = 6.06GTITGTLSADD1213 pKa = 3.02EE1214 pKa = 4.63HH1215 pKa = 6.66AQISIDD1221 pKa = 5.35NGVTWINLAIVDD1233 pKa = 4.54GKK1235 pKa = 9.89WSYY1238 pKa = 11.86NEE1240 pKa = 3.69TRR1242 pKa = 11.84TSVDD1246 pKa = 4.82GIDD1249 pKa = 3.17QTYY1252 pKa = 7.95TYY1254 pKa = 9.72QVRR1257 pKa = 11.84VVDD1260 pKa = 3.57NAGNVGSTDD1269 pKa = 3.43SQVVKK1274 pKa = 10.61ISTTVPDD1281 pKa = 3.36NTITVTTLLTADD1293 pKa = 3.69TTPTISGAYY1302 pKa = 9.71AKK1304 pKa = 10.74ALGNGEE1310 pKa = 4.36TIQVIVNGVTYY1321 pKa = 10.22KK1322 pKa = 10.71QGDD1325 pKa = 4.05GYY1327 pKa = 10.32LTVNTTAKK1335 pKa = 8.73TWSLHH1340 pKa = 6.08IPDD1343 pKa = 5.07ANALNVNGATDD1354 pKa = 3.59TPYY1357 pKa = 10.95SIIAQIVNIAGNVVSDD1373 pKa = 3.6TTANEE1378 pKa = 3.75LTVYY1382 pKa = 10.57QDD1384 pKa = 3.27VSISAGPSPDD1394 pKa = 3.72YY1395 pKa = 10.44MGQQPLFNGMTKK1407 pKa = 9.77IGAGEE1412 pKa = 3.85KK1413 pKa = 10.74LEE1415 pKa = 4.21VYY1417 pKa = 10.06VYY1419 pKa = 11.1NSTGTLIKK1427 pKa = 9.94TFSSVNGASVVDD1439 pKa = 3.8GLVFDD1444 pKa = 4.62ATTGSWKK1451 pKa = 8.42ITAANWGTSQLAGGSYY1467 pKa = 8.64TVQAKK1472 pKa = 9.26VVVADD1477 pKa = 4.28GSGGQISDD1485 pKa = 3.48VQAITVIQPIAITVPNSTYY1504 pKa = 11.03SDD1506 pKa = 3.51INSKK1510 pKa = 10.47VYY1512 pKa = 10.77ALDD1515 pKa = 4.15DD1516 pKa = 4.08GGYY1519 pKa = 10.67LLIWAQNTNNNTGSAASNYY1538 pKa = 10.36DD1539 pKa = 3.6LVAQRR1544 pKa = 11.84YY1545 pKa = 4.52TQSGVAVGSLIKK1557 pKa = 9.58ITNTTTGTYY1566 pKa = 10.64ANSEE1570 pKa = 4.36GYY1572 pKa = 10.79ADD1574 pKa = 5.53RR1575 pKa = 11.84NDD1577 pKa = 3.3MWDD1580 pKa = 3.44SNGSYY1585 pKa = 10.01SANVNGDD1592 pKa = 3.13GSFTIAYY1599 pKa = 9.42NYY1601 pKa = 11.04DD1602 pKa = 3.6LATQSAIKK1610 pKa = 10.82SFDD1613 pKa = 3.37DD1614 pKa = 4.03SGNLLSTTTVNSGQNYY1630 pKa = 7.84YY1631 pKa = 10.67FGYY1634 pKa = 10.7NYY1636 pKa = 10.25VEE1638 pKa = 4.19TTNGKK1643 pKa = 9.4VVVFTSGTIHH1653 pKa = 7.79DD1654 pKa = 4.4YY1655 pKa = 11.3DD1656 pKa = 3.67IFIAGSVKK1664 pKa = 10.45SYY1666 pKa = 10.67SASNLTNEE1674 pKa = 4.45TNGGNGYY1681 pKa = 10.03IYY1683 pKa = 10.38NQYY1686 pKa = 8.79ATPTGYY1692 pKa = 10.59SGFGNSTNIEE1702 pKa = 4.24SVSAVSLGGSLVLVQYY1718 pKa = 11.58ADD1720 pKa = 3.68YY1721 pKa = 9.66TSTDD1725 pKa = 3.12AYY1727 pKa = 7.74PTKK1730 pKa = 9.93EE1731 pKa = 3.56NVMIVDD1737 pKa = 3.79YY1738 pKa = 10.97SSTGVATVLKK1748 pKa = 10.34SAVANYY1754 pKa = 9.88YY1755 pKa = 11.02DD1756 pKa = 4.04NGWQIGACSFVLKK1769 pKa = 10.7EE1770 pKa = 4.17GGFVSLWAGNQDD1782 pKa = 3.32SATSLTNGGTMDD1794 pKa = 4.1GFNVYY1799 pKa = 8.35SRR1801 pKa = 11.84RR1802 pKa = 11.84FSYY1805 pKa = 10.82DD1806 pKa = 2.56VGANTITALDD1816 pKa = 3.8TTEE1819 pKa = 4.91KK1820 pKa = 10.23MVNTTMNGVNGVGYY1834 pKa = 8.1STMSTGHH1841 pKa = 6.22FSGCALAQGGYY1852 pKa = 8.6VVVWTKK1858 pKa = 10.18MLSNSLSEE1866 pKa = 4.88VYY1868 pKa = 10.33SQSYY1872 pKa = 9.55DD1873 pKa = 3.16AAGNRR1878 pKa = 11.84LGGEE1882 pKa = 4.48TLVSTQTVDD1891 pKa = 3.0GTGTIDD1897 pKa = 3.46TLPTVTALKK1906 pKa = 10.72DD1907 pKa = 3.19GGYY1910 pKa = 9.52VVSWTNSASTSYY1922 pKa = 10.87AINSTTTDD1930 pKa = 2.74IKK1932 pKa = 10.07TVIVNGDD1939 pKa = 3.34GTLRR1943 pKa = 11.84GADD1946 pKa = 3.47EE1947 pKa = 4.24THH1949 pKa = 7.0SIDD1952 pKa = 3.31ATYY1955 pKa = 11.36LNGTGTLIGTDD1966 pKa = 4.59GVCTLDD1972 pKa = 3.48GRR1974 pKa = 11.84NGATTMIGGKK1984 pKa = 10.22DD1985 pKa = 3.31NDD1987 pKa = 3.92NIIIKK1992 pKa = 9.89DD1993 pKa = 3.47INFSHH1998 pKa = 7.4IDD2000 pKa = 3.26GGAGNDD2006 pKa = 3.59TLIWDD2011 pKa = 4.14SSANLNLGDD2020 pKa = 5.39ILDD2023 pKa = 3.88KK2024 pKa = 10.81VQNIEE2029 pKa = 4.26TIHH2032 pKa = 7.7LGDD2035 pKa = 4.12ANANTMTVSIDD2046 pKa = 5.18DD2047 pKa = 3.92ILQMTSNNTLIIQGGDD2063 pKa = 3.08TDD2065 pKa = 3.75KK2066 pKa = 11.6VQIVDD2071 pKa = 3.66ANSWTSGGTQSYY2083 pKa = 10.32HH2084 pKa = 5.59GADD2087 pKa = 3.59YY2088 pKa = 10.89QIYY2091 pKa = 8.01TATTSDD2097 pKa = 2.99NHH2099 pKa = 7.29LINLWIQNNLGVIAA2113 pKa = 5.11
MM1 pKa = 7.37GKK3 pKa = 8.14VTATDD8 pKa = 3.82GAGNTGPEE16 pKa = 4.25ATTGAADD23 pKa = 3.4VTAPVAPTVDD33 pKa = 3.55QNNAAGLGGTTEE45 pKa = 4.19PNAVITVEE53 pKa = 4.36LGNGSTVTTTADD65 pKa = 3.42SQGNWLLEE73 pKa = 4.28PNPLAEE79 pKa = 4.35GEE81 pKa = 4.43VGKK84 pKa = 8.63VTATDD89 pKa = 3.8GAGNSSTPTMTDD101 pKa = 2.73GTDD104 pKa = 2.75ITAPVAPTVDD114 pKa = 3.55HH115 pKa = 6.82NNAAGLGGTTEE126 pKa = 4.19PNAVITVEE134 pKa = 4.36LGNGSTVTTTADD146 pKa = 3.48SQGNWNIKK154 pKa = 9.15PNPIAEE160 pKa = 4.5GEE162 pKa = 4.4SATVIATDD170 pKa = 3.52KK171 pKa = 11.23AGNTSAEE178 pKa = 4.36TPTGKK183 pKa = 10.45ADD185 pKa = 3.77TVAPTAPGIDD195 pKa = 3.55QNNEE199 pKa = 3.73SGLSGTAEE207 pKa = 4.14PEE209 pKa = 4.03STISVEE215 pKa = 4.14LADD218 pKa = 4.02GTTITTTTDD227 pKa = 2.92TAGNWSITPNPLGDD241 pKa = 3.81GEE243 pKa = 4.53VGKK246 pKa = 8.62VTATDD251 pKa = 3.38AAGNNSAPTMTDD263 pKa = 2.7GTDD266 pKa = 3.19ITPPAAPGVDD276 pKa = 3.42QNNGEE281 pKa = 4.36GLSGTAEE288 pKa = 3.99AGATVTVKK296 pKa = 10.73DD297 pKa = 3.6ADD299 pKa = 4.11GNTVTTTADD308 pKa = 3.54STGHH312 pKa = 5.99WSITPNPISDD322 pKa = 3.67GMSGTVTATDD332 pKa = 3.45EE333 pKa = 4.79AGNEE337 pKa = 4.35SNPSNTGTADD347 pKa = 3.29TVAPNAPLVEE357 pKa = 4.24QNNAAGLAGTTEE369 pKa = 4.29PNAVITVDD377 pKa = 4.15LGNGSTVTTMADD389 pKa = 3.57SQGHH393 pKa = 5.15WSIEE397 pKa = 3.99PNPLEE402 pKa = 4.11TGEE405 pKa = 4.25VGKK408 pKa = 8.62VTATDD413 pKa = 3.42AAGNTSTPTMTDD425 pKa = 2.87GTDD428 pKa = 3.19ITPPAAPGIDD438 pKa = 3.42QNNGEE443 pKa = 4.58GLSGTAEE450 pKa = 3.99AGATVTVKK458 pKa = 10.73DD459 pKa = 3.6ADD461 pKa = 4.11GNTVTTTADD470 pKa = 3.54STGHH474 pKa = 5.99WSITPNPIADD484 pKa = 3.95GTSGTVTATDD494 pKa = 3.41EE495 pKa = 4.64AGNEE499 pKa = 4.12SSPSNTGTADD509 pKa = 3.29TVAPNAPLVEE519 pKa = 4.24QNNAAGLAGTTEE531 pKa = 4.29PNAVITVDD539 pKa = 4.15LGNGSTVTTMADD551 pKa = 3.57SQGHH555 pKa = 5.15WSIEE559 pKa = 3.99PNPLEE564 pKa = 4.11TGEE567 pKa = 4.25VGKK570 pKa = 8.62VTATDD575 pKa = 3.42AAGNTSTPTMTDD587 pKa = 2.87GTDD590 pKa = 3.19ITPPAAPGVDD600 pKa = 3.42QNNGEE605 pKa = 4.36GLSGTAEE612 pKa = 3.99AGATVTVKK620 pKa = 10.73DD621 pKa = 3.6ADD623 pKa = 4.11GNTVTTTADD632 pKa = 3.54STGHH636 pKa = 5.99WSITPNPITDD646 pKa = 3.61GSEE649 pKa = 4.22GSVTATDD656 pKa = 3.58AAGNTSAEE664 pKa = 4.55TPTGTADD671 pKa = 3.51TVAPTAPGIDD681 pKa = 3.53QNNGEE686 pKa = 4.58GLSGTAEE693 pKa = 3.99AGATVTVKK701 pKa = 10.73DD702 pKa = 3.6ADD704 pKa = 4.11GNTVTTTADD713 pKa = 3.54STGHH717 pKa = 5.99WSITPNPITDD727 pKa = 3.86GTSGTVTATDD737 pKa = 3.41EE738 pKa = 4.64AGNEE742 pKa = 4.12SSPSNTGIADD752 pKa = 4.02TVAPTAPGIDD762 pKa = 3.53QNNGEE767 pKa = 4.41GLSGTVEE774 pKa = 3.88AGATVIVKK782 pKa = 10.21DD783 pKa = 3.6ADD785 pKa = 3.83GNTVTTTADD794 pKa = 3.54STGHH798 pKa = 5.99WSITPNPLADD808 pKa = 3.81GSEE811 pKa = 4.35GSVTATDD818 pKa = 3.58AAGNTSAEE826 pKa = 4.55TPTGTADD833 pKa = 3.51TVAPTAPGIDD843 pKa = 3.53QNNGEE848 pKa = 4.58GLSGTAEE855 pKa = 3.99AGATVTVKK863 pKa = 10.73DD864 pKa = 3.6ADD866 pKa = 4.11GNTVTTTADD875 pKa = 3.54STGHH879 pKa = 5.99WSITPNPITDD889 pKa = 3.86GTSGTVTATDD899 pKa = 3.41EE900 pKa = 4.64AGNEE904 pKa = 4.05SSPTNTGIADD914 pKa = 4.18TVAPTAPGIDD924 pKa = 3.53QNNGEE929 pKa = 4.63GLTGTAEE936 pKa = 4.15AGSTVIVKK944 pKa = 10.26DD945 pKa = 3.34ADD947 pKa = 3.83GNTVTTTADD956 pKa = 3.54STGHH960 pKa = 5.99WSITPNPITDD970 pKa = 3.61GSEE973 pKa = 4.22GSVTATDD980 pKa = 3.58AAGNTSAEE988 pKa = 4.55TPTGTADD995 pKa = 4.48LVAPDD1000 pKa = 4.27KK1001 pKa = 11.15PLVNLVNGIDD1011 pKa = 4.65PITGTAEE1018 pKa = 3.88PNSIVTVTYY1027 pKa = 11.07LDD1029 pKa = 3.91GTSVTVTASSDD1040 pKa = 3.76GSWSVLNPGLEE1051 pKa = 4.58DD1052 pKa = 4.31GDD1054 pKa = 4.71TITVTATDD1062 pKa = 3.59EE1063 pKa = 4.24VGNKK1067 pKa = 9.61SEE1069 pKa = 4.43SVTVTVDD1076 pKa = 3.12TYY1078 pKa = 11.72VLADD1082 pKa = 3.45TVIITNGVTNDD1093 pKa = 3.57DD1094 pKa = 3.61TPAIVGTAEE1103 pKa = 3.99ANSIVTLTIIGTTGSQVVSVSVDD1126 pKa = 3.18GTGVWTYY1133 pKa = 10.24TPTVALAEE1141 pKa = 4.09DD1142 pKa = 5.12VYY1144 pKa = 10.53TLSAIARR1151 pKa = 11.84DD1152 pKa = 3.56SEE1154 pKa = 4.26GRR1156 pKa = 11.84VSQIGASSITVDD1168 pKa = 3.29TTPPSSEE1175 pKa = 3.47IVIEE1179 pKa = 4.02ITAITDD1185 pKa = 3.39DD1186 pKa = 4.21SGVLGDD1192 pKa = 5.9FITNDD1197 pKa = 3.61TTPTIHH1203 pKa = 6.06GTITGTLSADD1213 pKa = 3.02EE1214 pKa = 4.63HH1215 pKa = 6.66AQISIDD1221 pKa = 5.35NGVTWINLAIVDD1233 pKa = 4.54GKK1235 pKa = 9.89WSYY1238 pKa = 11.86NEE1240 pKa = 3.69TRR1242 pKa = 11.84TSVDD1246 pKa = 4.82GIDD1249 pKa = 3.17QTYY1252 pKa = 7.95TYY1254 pKa = 9.72QVRR1257 pKa = 11.84VVDD1260 pKa = 3.57NAGNVGSTDD1269 pKa = 3.43SQVVKK1274 pKa = 10.61ISTTVPDD1281 pKa = 3.36NTITVTTLLTADD1293 pKa = 3.69TTPTISGAYY1302 pKa = 9.71AKK1304 pKa = 10.74ALGNGEE1310 pKa = 4.36TIQVIVNGVTYY1321 pKa = 10.22KK1322 pKa = 10.71QGDD1325 pKa = 4.05GYY1327 pKa = 10.32LTVNTTAKK1335 pKa = 8.73TWSLHH1340 pKa = 6.08IPDD1343 pKa = 5.07ANALNVNGATDD1354 pKa = 3.59TPYY1357 pKa = 10.95SIIAQIVNIAGNVVSDD1373 pKa = 3.6TTANEE1378 pKa = 3.75LTVYY1382 pKa = 10.57QDD1384 pKa = 3.27VSISAGPSPDD1394 pKa = 3.72YY1395 pKa = 10.44MGQQPLFNGMTKK1407 pKa = 9.77IGAGEE1412 pKa = 3.85KK1413 pKa = 10.74LEE1415 pKa = 4.21VYY1417 pKa = 10.06VYY1419 pKa = 11.1NSTGTLIKK1427 pKa = 9.94TFSSVNGASVVDD1439 pKa = 3.8GLVFDD1444 pKa = 4.62ATTGSWKK1451 pKa = 8.42ITAANWGTSQLAGGSYY1467 pKa = 8.64TVQAKK1472 pKa = 9.26VVVADD1477 pKa = 4.28GSGGQISDD1485 pKa = 3.48VQAITVIQPIAITVPNSTYY1504 pKa = 11.03SDD1506 pKa = 3.51INSKK1510 pKa = 10.47VYY1512 pKa = 10.77ALDD1515 pKa = 4.15DD1516 pKa = 4.08GGYY1519 pKa = 10.67LLIWAQNTNNNTGSAASNYY1538 pKa = 10.36DD1539 pKa = 3.6LVAQRR1544 pKa = 11.84YY1545 pKa = 4.52TQSGVAVGSLIKK1557 pKa = 9.58ITNTTTGTYY1566 pKa = 10.64ANSEE1570 pKa = 4.36GYY1572 pKa = 10.79ADD1574 pKa = 5.53RR1575 pKa = 11.84NDD1577 pKa = 3.3MWDD1580 pKa = 3.44SNGSYY1585 pKa = 10.01SANVNGDD1592 pKa = 3.13GSFTIAYY1599 pKa = 9.42NYY1601 pKa = 11.04DD1602 pKa = 3.6LATQSAIKK1610 pKa = 10.82SFDD1613 pKa = 3.37DD1614 pKa = 4.03SGNLLSTTTVNSGQNYY1630 pKa = 7.84YY1631 pKa = 10.67FGYY1634 pKa = 10.7NYY1636 pKa = 10.25VEE1638 pKa = 4.19TTNGKK1643 pKa = 9.4VVVFTSGTIHH1653 pKa = 7.79DD1654 pKa = 4.4YY1655 pKa = 11.3DD1656 pKa = 3.67IFIAGSVKK1664 pKa = 10.45SYY1666 pKa = 10.67SASNLTNEE1674 pKa = 4.45TNGGNGYY1681 pKa = 10.03IYY1683 pKa = 10.38NQYY1686 pKa = 8.79ATPTGYY1692 pKa = 10.59SGFGNSTNIEE1702 pKa = 4.24SVSAVSLGGSLVLVQYY1718 pKa = 11.58ADD1720 pKa = 3.68YY1721 pKa = 9.66TSTDD1725 pKa = 3.12AYY1727 pKa = 7.74PTKK1730 pKa = 9.93EE1731 pKa = 3.56NVMIVDD1737 pKa = 3.79YY1738 pKa = 10.97SSTGVATVLKK1748 pKa = 10.34SAVANYY1754 pKa = 9.88YY1755 pKa = 11.02DD1756 pKa = 4.04NGWQIGACSFVLKK1769 pKa = 10.7EE1770 pKa = 4.17GGFVSLWAGNQDD1782 pKa = 3.32SATSLTNGGTMDD1794 pKa = 4.1GFNVYY1799 pKa = 8.35SRR1801 pKa = 11.84RR1802 pKa = 11.84FSYY1805 pKa = 10.82DD1806 pKa = 2.56VGANTITALDD1816 pKa = 3.8TTEE1819 pKa = 4.91KK1820 pKa = 10.23MVNTTMNGVNGVGYY1834 pKa = 8.1STMSTGHH1841 pKa = 6.22FSGCALAQGGYY1852 pKa = 8.6VVVWTKK1858 pKa = 10.18MLSNSLSEE1866 pKa = 4.88VYY1868 pKa = 10.33SQSYY1872 pKa = 9.55DD1873 pKa = 3.16AAGNRR1878 pKa = 11.84LGGEE1882 pKa = 4.48TLVSTQTVDD1891 pKa = 3.0GTGTIDD1897 pKa = 3.46TLPTVTALKK1906 pKa = 10.72DD1907 pKa = 3.19GGYY1910 pKa = 9.52VVSWTNSASTSYY1922 pKa = 10.87AINSTTTDD1930 pKa = 2.74IKK1932 pKa = 10.07TVIVNGDD1939 pKa = 3.34GTLRR1943 pKa = 11.84GADD1946 pKa = 3.47EE1947 pKa = 4.24THH1949 pKa = 7.0SIDD1952 pKa = 3.31ATYY1955 pKa = 11.36LNGTGTLIGTDD1966 pKa = 4.59GVCTLDD1972 pKa = 3.48GRR1974 pKa = 11.84NGATTMIGGKK1984 pKa = 10.22DD1985 pKa = 3.31NDD1987 pKa = 3.92NIIIKK1992 pKa = 9.89DD1993 pKa = 3.47INFSHH1998 pKa = 7.4IDD2000 pKa = 3.26GGAGNDD2006 pKa = 3.59TLIWDD2011 pKa = 4.14SSANLNLGDD2020 pKa = 5.39ILDD2023 pKa = 3.88KK2024 pKa = 10.81VQNIEE2029 pKa = 4.26TIHH2032 pKa = 7.7LGDD2035 pKa = 4.12ANANTMTVSIDD2046 pKa = 5.18DD2047 pKa = 3.92ILQMTSNNTLIIQGGDD2063 pKa = 3.08TDD2065 pKa = 3.75KK2066 pKa = 11.6VQIVDD2071 pKa = 3.66ANSWTSGGTQSYY2083 pKa = 10.32HH2084 pKa = 5.59GADD2087 pKa = 3.59YY2088 pKa = 10.89QIYY2091 pKa = 8.01TATTSDD2097 pKa = 2.99NHH2099 pKa = 7.29LINLWIQNNLGVIAA2113 pKa = 5.11
Molecular weight: 213.31 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2U3N3H8|A0A2U3N3H8_9GAMM Elongation factor G OS=Acinetobacter stercoris OX=2126983 GN=fusA PE=3 SV=1
MM1 pKa = 7.31IQTLTAIQTPTATQTRR17 pKa = 11.84TAXQTRR23 pKa = 11.84TAIQTXTATQTXTXIQTPTATQTPTVIQTXTAIQTXTATQTRR65 pKa = 11.84TVIQTLTAIQTPTXTQTLTAXQXRR89 pKa = 11.84TAIQTLTATQTXTXXQTXTXIQTLTAIQTPTATQTPTVIQTLTAIQTPTXTQTRR143 pKa = 11.84TXIQTPTXTQTRR155 pKa = 11.84TATQTPTVIQTLTATQTLTATQTLTVTQTPTLMNRR190 pKa = 11.84MILHH194 pKa = 6.52LMKK197 pKa = 9.99MVQNLQVQQKK207 pKa = 10.14LALQLLL213 pKa = 4.27
MM1 pKa = 7.31IQTLTAIQTPTATQTRR17 pKa = 11.84TAXQTRR23 pKa = 11.84TAIQTXTATQTXTXIQTPTATQTPTVIQTXTAIQTXTATQTRR65 pKa = 11.84TVIQTLTAIQTPTXTQTLTAXQXRR89 pKa = 11.84TAIQTLTATQTXTXXQTXTXIQTLTAIQTPTATQTPTVIQTLTAIQTPTXTQTRR143 pKa = 11.84TXIQTPTXTQTRR155 pKa = 11.84TATQTPTVIQTLTATQTLTATQTLTVTQTPTLMNRR190 pKa = 11.84MILHH194 pKa = 6.52LMKK197 pKa = 9.99MVQNLQVQQKK207 pKa = 10.14LALQLLL213 pKa = 4.27
Molecular weight: 21.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1176768 |
30 |
4370 |
305.1 |
34.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.103 ± 0.049 | 0.923 ± 0.013 |
5.332 ± 0.042 | 5.783 ± 0.038 |
4.384 ± 0.034 | 6.529 ± 0.047 |
2.373 ± 0.023 | 7.314 ± 0.031 |
5.969 ± 0.038 | 10.172 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.405 ± 0.023 | 4.805 ± 0.041 |
3.891 ± 0.028 | 5.06 ± 0.041 |
4.097 ± 0.033 | 6.367 ± 0.036 |
5.385 ± 0.045 | 6.473 ± 0.033 |
1.261 ± 0.017 | 3.368 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
