
Alternaria alternata chrysovirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Chrysoviridae; Betachrysovirus; Alternaria alternata chrysovirus
Average proteome isoelectric point is 6.66
Get precalculated fractions of proteins
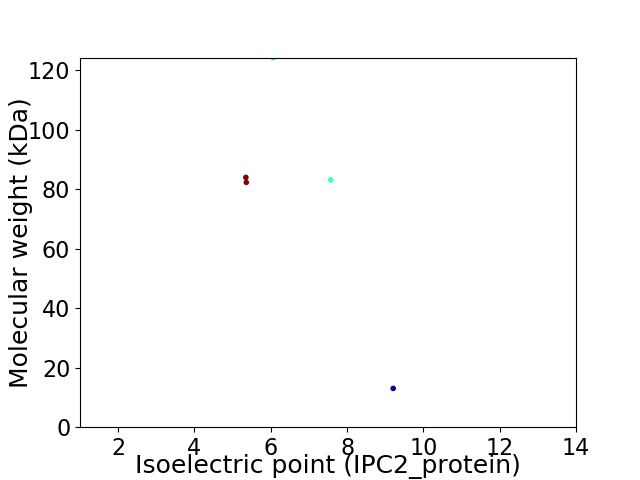
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z5X7B8|A0A2Z5X7B8_9VIRU Uncharacterized protein OS=Alternaria alternata chrysovirus 1 OX=2066695 PE=4 SV=1
MM1 pKa = 7.96ADD3 pKa = 3.24QSEE6 pKa = 4.8DD7 pKa = 3.75LPPSYY12 pKa = 11.15SLTPNTTSSSPSARR26 pKa = 11.84PFEE29 pKa = 4.61RR30 pKa = 11.84MMEE33 pKa = 4.16ALTVRR38 pKa = 11.84SAAVTTWSEE47 pKa = 3.95QVEE50 pKa = 4.37AEE52 pKa = 4.24AEE54 pKa = 3.92EE55 pKa = 4.9GLQQEE60 pKa = 4.72VSVGGTSLKK69 pKa = 10.37LARR72 pKa = 11.84LDD74 pKa = 3.35YY75 pKa = 11.36GMARR79 pKa = 11.84DD80 pKa = 3.77PVPPQEE86 pKa = 3.83RR87 pKa = 11.84LQYY90 pKa = 10.53QRR92 pKa = 11.84GLRR95 pKa = 11.84GTTLLRR101 pKa = 11.84TAASRR106 pKa = 11.84AAVPCSDD113 pKa = 4.08CEE115 pKa = 3.91ISRR118 pKa = 11.84VCAGVPYY125 pKa = 10.39GPGVKK130 pKa = 10.13SLGWTKK136 pKa = 11.04AEE138 pKa = 4.06QKK140 pKa = 10.9ASLVVLDD147 pKa = 5.23TVASMLVRR155 pKa = 11.84GFRR158 pKa = 11.84KK159 pKa = 9.71GVSKK163 pKa = 8.32MTLPKK168 pKa = 10.26GSRR171 pKa = 11.84VTLSEE176 pKa = 3.84WPEE179 pKa = 3.52QNLYY183 pKa = 8.24HH184 pKa = 6.31TEE186 pKa = 3.69VGYY189 pKa = 7.82TWWSAVLVAPSDD201 pKa = 3.75NIIRR205 pKa = 11.84VMMHH209 pKa = 5.95VDD211 pKa = 3.38GTTLLDD217 pKa = 3.24IAGGDD222 pKa = 3.47EE223 pKa = 4.09MLEE226 pKa = 4.08ARR228 pKa = 11.84HH229 pKa = 6.42RR230 pKa = 11.84DD231 pKa = 3.57PDD233 pKa = 3.7LHH235 pKa = 7.88SDD237 pKa = 4.37LLVLLAGVQHH247 pKa = 7.34AILADD252 pKa = 3.76EE253 pKa = 5.02LSASYY258 pKa = 10.71HH259 pKa = 5.97LLQDD263 pKa = 3.53ASLATMRR270 pKa = 11.84QHH272 pKa = 7.28VSTTSAGSTDD282 pKa = 3.18QGGYY286 pKa = 10.67AGGGPNHH293 pKa = 6.68ALWSEE298 pKa = 4.06GEE300 pKa = 4.23YY301 pKa = 10.98LSDD304 pKa = 5.59DD305 pKa = 3.65LVQDD309 pKa = 3.41RR310 pKa = 11.84RR311 pKa = 11.84VPTAYY316 pKa = 10.78YY317 pKa = 10.24DD318 pKa = 3.76ANLDD322 pKa = 3.47LTAYY326 pKa = 9.7GLASAGDD333 pKa = 4.09LPPAVLVALGANGGKK348 pKa = 10.28SITGSVLTLRR358 pKa = 11.84NSSAAGGFFLAVTSATANAKK378 pKa = 9.93PYY380 pKa = 10.84ALEE383 pKa = 4.37KK384 pKa = 10.52SLKK387 pKa = 10.45LGATVRR393 pKa = 11.84DD394 pKa = 4.0GLFSPACVGALCSRR408 pKa = 11.84APGLVGVVVDD418 pKa = 3.8NEE420 pKa = 4.45YY421 pKa = 10.73IAMQTVLVRR430 pKa = 11.84LDD432 pKa = 3.78PEE434 pKa = 4.9DD435 pKa = 4.24PLVLYY440 pKa = 10.76DD441 pKa = 3.84RR442 pKa = 11.84DD443 pKa = 3.64PYY445 pKa = 11.14RR446 pKa = 11.84LYY448 pKa = 10.74EE449 pKa = 4.19YY450 pKa = 10.98VWDD453 pKa = 3.96LCGVMGWRR461 pKa = 11.84IEE463 pKa = 4.03AAAGYY468 pKa = 10.73AMEE471 pKa = 5.06VYY473 pKa = 10.71SGDD476 pKa = 3.38TRR478 pKa = 11.84LGDD481 pKa = 3.31VAVRR485 pKa = 11.84YY486 pKa = 8.63GAAVGPRR493 pKa = 11.84WLRR496 pKa = 11.84LLTLAAIEE504 pKa = 4.09PRR506 pKa = 11.84KK507 pKa = 10.24SRR509 pKa = 11.84WSPEE513 pKa = 3.61LLLDD517 pKa = 3.83YY518 pKa = 10.41GASGLGRR525 pKa = 11.84YY526 pKa = 8.07LLRR529 pKa = 11.84EE530 pKa = 4.15YY531 pKa = 10.64ILAGRR536 pKa = 11.84FAGFKK541 pKa = 10.14SLMMHH546 pKa = 5.65TEE548 pKa = 4.54GYY550 pKa = 9.29FAEE553 pKa = 4.41AAKK556 pKa = 10.59SIASPKK562 pKa = 9.44PNAPAAKK569 pKa = 9.95LPVSYY574 pKa = 11.06VIDD577 pKa = 3.61TEE579 pKa = 4.23YY580 pKa = 10.28LTRR583 pKa = 11.84KK584 pKa = 9.9LPDD587 pKa = 3.32GSIEE591 pKa = 4.16RR592 pKa = 11.84FVYY595 pKa = 10.75ALGLAKK601 pKa = 9.95FYY603 pKa = 10.97AGNYY607 pKa = 8.2VGSALVVDD615 pKa = 4.59QSPEE619 pKa = 3.96LEE621 pKa = 4.77QFIQDD626 pKa = 3.86NEE628 pKa = 4.28HH629 pKa = 6.54LAGTKK634 pKa = 9.33PMRR637 pKa = 11.84MLASIRR643 pKa = 11.84EE644 pKa = 4.25EE645 pKa = 3.72IAPRR649 pKa = 11.84ASLCNGNPSALLAEE663 pKa = 4.41LRR665 pKa = 11.84IAARR669 pKa = 11.84SPEE672 pKa = 3.65VRR674 pKa = 11.84LYY676 pKa = 11.36AKK678 pKa = 10.46GADD681 pKa = 3.69AEE683 pKa = 4.41RR684 pKa = 11.84EE685 pKa = 4.14LLSSKK690 pKa = 10.11LARR693 pKa = 11.84GTRR696 pKa = 11.84LFRR699 pKa = 11.84RR700 pKa = 11.84ANLAAPLRR708 pKa = 11.84VPLHH712 pKa = 6.18EE713 pKa = 5.35LGAMVPRR720 pKa = 11.84YY721 pKa = 9.85EE722 pKa = 4.04EE723 pKa = 3.94LAAKK727 pKa = 9.95HH728 pKa = 6.49DD729 pKa = 4.39WDD731 pKa = 4.86PSHH734 pKa = 6.81NPAKK738 pKa = 10.21EE739 pKa = 3.88CVLFGIEE746 pKa = 4.71AGLGDD751 pKa = 4.53ALPTEE756 pKa = 4.5RR757 pKa = 11.84AVGLDD762 pKa = 3.65DD763 pKa = 5.14MYY765 pKa = 11.58TLLPQMSTQVVV776 pKa = 3.27
MM1 pKa = 7.96ADD3 pKa = 3.24QSEE6 pKa = 4.8DD7 pKa = 3.75LPPSYY12 pKa = 11.15SLTPNTTSSSPSARR26 pKa = 11.84PFEE29 pKa = 4.61RR30 pKa = 11.84MMEE33 pKa = 4.16ALTVRR38 pKa = 11.84SAAVTTWSEE47 pKa = 3.95QVEE50 pKa = 4.37AEE52 pKa = 4.24AEE54 pKa = 3.92EE55 pKa = 4.9GLQQEE60 pKa = 4.72VSVGGTSLKK69 pKa = 10.37LARR72 pKa = 11.84LDD74 pKa = 3.35YY75 pKa = 11.36GMARR79 pKa = 11.84DD80 pKa = 3.77PVPPQEE86 pKa = 3.83RR87 pKa = 11.84LQYY90 pKa = 10.53QRR92 pKa = 11.84GLRR95 pKa = 11.84GTTLLRR101 pKa = 11.84TAASRR106 pKa = 11.84AAVPCSDD113 pKa = 4.08CEE115 pKa = 3.91ISRR118 pKa = 11.84VCAGVPYY125 pKa = 10.39GPGVKK130 pKa = 10.13SLGWTKK136 pKa = 11.04AEE138 pKa = 4.06QKK140 pKa = 10.9ASLVVLDD147 pKa = 5.23TVASMLVRR155 pKa = 11.84GFRR158 pKa = 11.84KK159 pKa = 9.71GVSKK163 pKa = 8.32MTLPKK168 pKa = 10.26GSRR171 pKa = 11.84VTLSEE176 pKa = 3.84WPEE179 pKa = 3.52QNLYY183 pKa = 8.24HH184 pKa = 6.31TEE186 pKa = 3.69VGYY189 pKa = 7.82TWWSAVLVAPSDD201 pKa = 3.75NIIRR205 pKa = 11.84VMMHH209 pKa = 5.95VDD211 pKa = 3.38GTTLLDD217 pKa = 3.24IAGGDD222 pKa = 3.47EE223 pKa = 4.09MLEE226 pKa = 4.08ARR228 pKa = 11.84HH229 pKa = 6.42RR230 pKa = 11.84DD231 pKa = 3.57PDD233 pKa = 3.7LHH235 pKa = 7.88SDD237 pKa = 4.37LLVLLAGVQHH247 pKa = 7.34AILADD252 pKa = 3.76EE253 pKa = 5.02LSASYY258 pKa = 10.71HH259 pKa = 5.97LLQDD263 pKa = 3.53ASLATMRR270 pKa = 11.84QHH272 pKa = 7.28VSTTSAGSTDD282 pKa = 3.18QGGYY286 pKa = 10.67AGGGPNHH293 pKa = 6.68ALWSEE298 pKa = 4.06GEE300 pKa = 4.23YY301 pKa = 10.98LSDD304 pKa = 5.59DD305 pKa = 3.65LVQDD309 pKa = 3.41RR310 pKa = 11.84RR311 pKa = 11.84VPTAYY316 pKa = 10.78YY317 pKa = 10.24DD318 pKa = 3.76ANLDD322 pKa = 3.47LTAYY326 pKa = 9.7GLASAGDD333 pKa = 4.09LPPAVLVALGANGGKK348 pKa = 10.28SITGSVLTLRR358 pKa = 11.84NSSAAGGFFLAVTSATANAKK378 pKa = 9.93PYY380 pKa = 10.84ALEE383 pKa = 4.37KK384 pKa = 10.52SLKK387 pKa = 10.45LGATVRR393 pKa = 11.84DD394 pKa = 4.0GLFSPACVGALCSRR408 pKa = 11.84APGLVGVVVDD418 pKa = 3.8NEE420 pKa = 4.45YY421 pKa = 10.73IAMQTVLVRR430 pKa = 11.84LDD432 pKa = 3.78PEE434 pKa = 4.9DD435 pKa = 4.24PLVLYY440 pKa = 10.76DD441 pKa = 3.84RR442 pKa = 11.84DD443 pKa = 3.64PYY445 pKa = 11.14RR446 pKa = 11.84LYY448 pKa = 10.74EE449 pKa = 4.19YY450 pKa = 10.98VWDD453 pKa = 3.96LCGVMGWRR461 pKa = 11.84IEE463 pKa = 4.03AAAGYY468 pKa = 10.73AMEE471 pKa = 5.06VYY473 pKa = 10.71SGDD476 pKa = 3.38TRR478 pKa = 11.84LGDD481 pKa = 3.31VAVRR485 pKa = 11.84YY486 pKa = 8.63GAAVGPRR493 pKa = 11.84WLRR496 pKa = 11.84LLTLAAIEE504 pKa = 4.09PRR506 pKa = 11.84KK507 pKa = 10.24SRR509 pKa = 11.84WSPEE513 pKa = 3.61LLLDD517 pKa = 3.83YY518 pKa = 10.41GASGLGRR525 pKa = 11.84YY526 pKa = 8.07LLRR529 pKa = 11.84EE530 pKa = 4.15YY531 pKa = 10.64ILAGRR536 pKa = 11.84FAGFKK541 pKa = 10.14SLMMHH546 pKa = 5.65TEE548 pKa = 4.54GYY550 pKa = 9.29FAEE553 pKa = 4.41AAKK556 pKa = 10.59SIASPKK562 pKa = 9.44PNAPAAKK569 pKa = 9.95LPVSYY574 pKa = 11.06VIDD577 pKa = 3.61TEE579 pKa = 4.23YY580 pKa = 10.28LTRR583 pKa = 11.84KK584 pKa = 9.9LPDD587 pKa = 3.32GSIEE591 pKa = 4.16RR592 pKa = 11.84FVYY595 pKa = 10.75ALGLAKK601 pKa = 9.95FYY603 pKa = 10.97AGNYY607 pKa = 8.2VGSALVVDD615 pKa = 4.59QSPEE619 pKa = 3.96LEE621 pKa = 4.77QFIQDD626 pKa = 3.86NEE628 pKa = 4.28HH629 pKa = 6.54LAGTKK634 pKa = 9.33PMRR637 pKa = 11.84MLASIRR643 pKa = 11.84EE644 pKa = 4.25EE645 pKa = 3.72IAPRR649 pKa = 11.84ASLCNGNPSALLAEE663 pKa = 4.41LRR665 pKa = 11.84IAARR669 pKa = 11.84SPEE672 pKa = 3.65VRR674 pKa = 11.84LYY676 pKa = 11.36AKK678 pKa = 10.46GADD681 pKa = 3.69AEE683 pKa = 4.41RR684 pKa = 11.84EE685 pKa = 4.14LLSSKK690 pKa = 10.11LARR693 pKa = 11.84GTRR696 pKa = 11.84LFRR699 pKa = 11.84RR700 pKa = 11.84ANLAAPLRR708 pKa = 11.84VPLHH712 pKa = 6.18EE713 pKa = 5.35LGAMVPRR720 pKa = 11.84YY721 pKa = 9.85EE722 pKa = 4.04EE723 pKa = 3.94LAAKK727 pKa = 9.95HH728 pKa = 6.49DD729 pKa = 4.39WDD731 pKa = 4.86PSHH734 pKa = 6.81NPAKK738 pKa = 10.21EE739 pKa = 3.88CVLFGIEE746 pKa = 4.71AGLGDD751 pKa = 4.53ALPTEE756 pKa = 4.5RR757 pKa = 11.84AVGLDD762 pKa = 3.65DD763 pKa = 5.14MYY765 pKa = 11.58TLLPQMSTQVVV776 pKa = 3.27
Molecular weight: 83.92 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z5X7J9|A0A2Z5X7J9_9VIRU Uncharacterized protein OS=Alternaria alternata chrysovirus 1 OX=2066695 PE=4 SV=1
MM1 pKa = 7.05MFWNKK6 pKa = 9.24RR7 pKa = 11.84QAAPGVMGSEE17 pKa = 4.07RR18 pKa = 11.84MKK20 pKa = 10.42VAGLIRR26 pKa = 11.84PATAVSTVPWWFLDD40 pKa = 3.63TPLRR44 pKa = 11.84TEE46 pKa = 4.32ASALCLLLTIWTHH59 pKa = 6.68LRR61 pKa = 11.84GACLAICDD69 pKa = 4.17RR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84SSDD75 pKa = 3.1QCSEE79 pKa = 3.73CRR81 pKa = 11.84ACVARR86 pKa = 11.84DD87 pKa = 3.19WRR89 pKa = 11.84HH90 pKa = 4.15TAWRR94 pKa = 11.84HH95 pKa = 4.95LRR97 pKa = 11.84WSLVAAGRR105 pKa = 11.84PDD107 pKa = 3.4ALRR110 pKa = 11.84GVNFGG115 pKa = 3.81
MM1 pKa = 7.05MFWNKK6 pKa = 9.24RR7 pKa = 11.84QAAPGVMGSEE17 pKa = 4.07RR18 pKa = 11.84MKK20 pKa = 10.42VAGLIRR26 pKa = 11.84PATAVSTVPWWFLDD40 pKa = 3.63TPLRR44 pKa = 11.84TEE46 pKa = 4.32ASALCLLLTIWTHH59 pKa = 6.68LRR61 pKa = 11.84GACLAICDD69 pKa = 4.17RR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84SSDD75 pKa = 3.1QCSEE79 pKa = 3.73CRR81 pKa = 11.84ACVARR86 pKa = 11.84DD87 pKa = 3.19WRR89 pKa = 11.84HH90 pKa = 4.15TAWRR94 pKa = 11.84HH95 pKa = 4.95LRR97 pKa = 11.84WSLVAAGRR105 pKa = 11.84PDD107 pKa = 3.4ALRR110 pKa = 11.84GVNFGG115 pKa = 3.81
Molecular weight: 13.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3553 |
115 |
1117 |
710.6 |
77.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.468 ± 0.262 | 1.407 ± 0.219 |
4.7 ± 0.339 | 6.445 ± 0.552 |
2.533 ± 0.422 | 7.543 ± 0.613 |
2.336 ± 0.313 | 2.252 ± 0.054 |
3.096 ± 0.457 | 10.667 ± 0.698 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.589 ± 0.108 | 2.083 ± 0.262 |
5.545 ± 0.817 | 3.8 ± 0.405 |
7.149 ± 0.303 | 7.656 ± 0.433 |
4.869 ± 0.314 | 8.19 ± 0.317 |
1.52 ± 0.261 | 3.152 ± 0.419 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
