
Haloquadratum walsbyi (strain DSM 16790 / HBSQ001)
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Haloferacales; Haloferacaceae; Haloquadratum; Haloquadratum walsbyi
Average proteome isoelectric point is 4.95
Get precalculated fractions of proteins
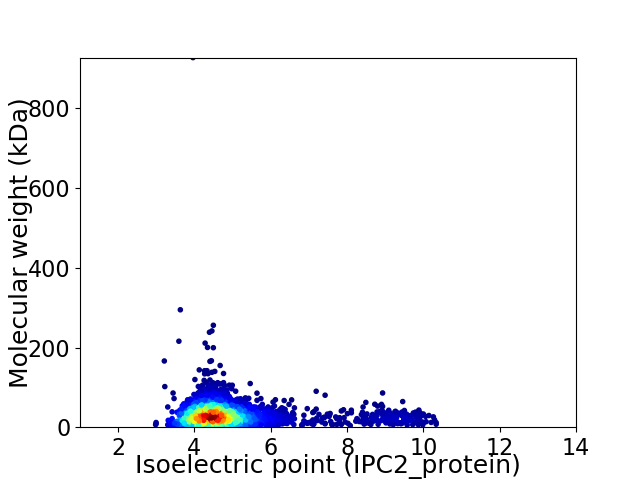
Virtual 2D-PAGE plot for 2558 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q18E23|Q18E23_HALWD Probable membrane transporter protein OS=Haloquadratum walsbyi (strain DSM 16790 / HBSQ001) OX=362976 GN=HQ_3724A PE=3 SV=1
MM1 pKa = 6.86TTDD4 pKa = 5.21DD5 pKa = 3.75YY6 pKa = 12.07AKK8 pKa = 10.53DD9 pKa = 3.65VLVSPEE15 pKa = 4.02WVTDD19 pKa = 3.64RR20 pKa = 11.84LDD22 pKa = 4.48EE23 pKa = 4.37IQSDD27 pKa = 3.91DD28 pKa = 3.52PDD30 pKa = 3.67YY31 pKa = 11.39RR32 pKa = 11.84LVEE35 pKa = 3.99VDD37 pKa = 4.33VDD39 pKa = 3.75TEE41 pKa = 4.55AYY43 pKa = 10.35DD44 pKa = 3.53EE45 pKa = 4.18AHH47 pKa = 6.81APGAIGFNWEE57 pKa = 4.25TQLQDD62 pKa = 3.16PTTRR66 pKa = 11.84DD67 pKa = 3.23ILDD70 pKa = 3.83KK71 pKa = 11.32EE72 pKa = 4.33DD73 pKa = 4.8FEE75 pKa = 4.87TLLGNHH81 pKa = 7.56GISDD85 pKa = 4.28DD86 pKa = 3.9STVIIYY92 pKa = 10.65GDD94 pKa = 3.44NSNWFASYY102 pKa = 9.16TYY104 pKa = 9.6WQFKK108 pKa = 10.46YY109 pKa = 10.45YY110 pKa = 10.89GHH112 pKa = 7.87DD113 pKa = 3.89DD114 pKa = 3.4VRR116 pKa = 11.84LLNGARR122 pKa = 11.84DD123 pKa = 3.57YY124 pKa = 11.05WLDD127 pKa = 3.03NDD129 pKa = 4.16YY130 pKa = 10.94PVTDD134 pKa = 4.23EE135 pKa = 4.06VPEE138 pKa = 4.14YY139 pKa = 10.59STVEE143 pKa = 3.89YY144 pKa = 9.8EE145 pKa = 3.8ARR147 pKa = 11.84GPFEE151 pKa = 4.77SIRR154 pKa = 11.84AYY156 pKa = 10.72RR157 pKa = 11.84DD158 pKa = 3.77DD159 pKa = 3.78VDD161 pKa = 4.12HH162 pKa = 7.51AIDD165 pKa = 3.57QDD167 pKa = 4.05LPLVDD172 pKa = 3.5VRR174 pKa = 11.84SPEE177 pKa = 4.06EE178 pKa = 3.62FSGEE182 pKa = 3.6ILAPPGLNEE191 pKa = 3.53TAQRR195 pKa = 11.84GGHH198 pKa = 6.15IPGASNISWADD209 pKa = 3.47TVNDD213 pKa = 4.04DD214 pKa = 4.14GTFKK218 pKa = 10.79SAEE221 pKa = 4.14EE222 pKa = 4.35LEE224 pKa = 4.83SIYY227 pKa = 11.12ADD229 pKa = 3.58YY230 pKa = 11.09DD231 pKa = 3.33IDD233 pKa = 4.29GSEE236 pKa = 4.11TTVAYY241 pKa = 10.32CRR243 pKa = 11.84IGEE246 pKa = 4.16RR247 pKa = 11.84SSIAWFALHH256 pKa = 6.85EE257 pKa = 4.13LLGYY261 pKa = 10.7DD262 pKa = 3.91EE263 pKa = 4.55VVNYY267 pKa = 10.35DD268 pKa = 3.71GSWTEE273 pKa = 3.62WGNLVGAPIEE283 pKa = 4.34TGGADD288 pKa = 3.04
MM1 pKa = 6.86TTDD4 pKa = 5.21DD5 pKa = 3.75YY6 pKa = 12.07AKK8 pKa = 10.53DD9 pKa = 3.65VLVSPEE15 pKa = 4.02WVTDD19 pKa = 3.64RR20 pKa = 11.84LDD22 pKa = 4.48EE23 pKa = 4.37IQSDD27 pKa = 3.91DD28 pKa = 3.52PDD30 pKa = 3.67YY31 pKa = 11.39RR32 pKa = 11.84LVEE35 pKa = 3.99VDD37 pKa = 4.33VDD39 pKa = 3.75TEE41 pKa = 4.55AYY43 pKa = 10.35DD44 pKa = 3.53EE45 pKa = 4.18AHH47 pKa = 6.81APGAIGFNWEE57 pKa = 4.25TQLQDD62 pKa = 3.16PTTRR66 pKa = 11.84DD67 pKa = 3.23ILDD70 pKa = 3.83KK71 pKa = 11.32EE72 pKa = 4.33DD73 pKa = 4.8FEE75 pKa = 4.87TLLGNHH81 pKa = 7.56GISDD85 pKa = 4.28DD86 pKa = 3.9STVIIYY92 pKa = 10.65GDD94 pKa = 3.44NSNWFASYY102 pKa = 9.16TYY104 pKa = 9.6WQFKK108 pKa = 10.46YY109 pKa = 10.45YY110 pKa = 10.89GHH112 pKa = 7.87DD113 pKa = 3.89DD114 pKa = 3.4VRR116 pKa = 11.84LLNGARR122 pKa = 11.84DD123 pKa = 3.57YY124 pKa = 11.05WLDD127 pKa = 3.03NDD129 pKa = 4.16YY130 pKa = 10.94PVTDD134 pKa = 4.23EE135 pKa = 4.06VPEE138 pKa = 4.14YY139 pKa = 10.59STVEE143 pKa = 3.89YY144 pKa = 9.8EE145 pKa = 3.8ARR147 pKa = 11.84GPFEE151 pKa = 4.77SIRR154 pKa = 11.84AYY156 pKa = 10.72RR157 pKa = 11.84DD158 pKa = 3.77DD159 pKa = 3.78VDD161 pKa = 4.12HH162 pKa = 7.51AIDD165 pKa = 3.57QDD167 pKa = 4.05LPLVDD172 pKa = 3.5VRR174 pKa = 11.84SPEE177 pKa = 4.06EE178 pKa = 3.62FSGEE182 pKa = 3.6ILAPPGLNEE191 pKa = 3.53TAQRR195 pKa = 11.84GGHH198 pKa = 6.15IPGASNISWADD209 pKa = 3.47TVNDD213 pKa = 4.04DD214 pKa = 4.14GTFKK218 pKa = 10.79SAEE221 pKa = 4.14EE222 pKa = 4.35LEE224 pKa = 4.83SIYY227 pKa = 11.12ADD229 pKa = 3.58YY230 pKa = 11.09DD231 pKa = 3.33IDD233 pKa = 4.29GSEE236 pKa = 4.11TTVAYY241 pKa = 10.32CRR243 pKa = 11.84IGEE246 pKa = 4.16RR247 pKa = 11.84SSIAWFALHH256 pKa = 6.85EE257 pKa = 4.13LLGYY261 pKa = 10.7DD262 pKa = 3.91EE263 pKa = 4.55VVNYY267 pKa = 10.35DD268 pKa = 3.71GSWTEE273 pKa = 3.62WGNLVGAPIEE283 pKa = 4.34TGGADD288 pKa = 3.04
Molecular weight: 32.46 kDa
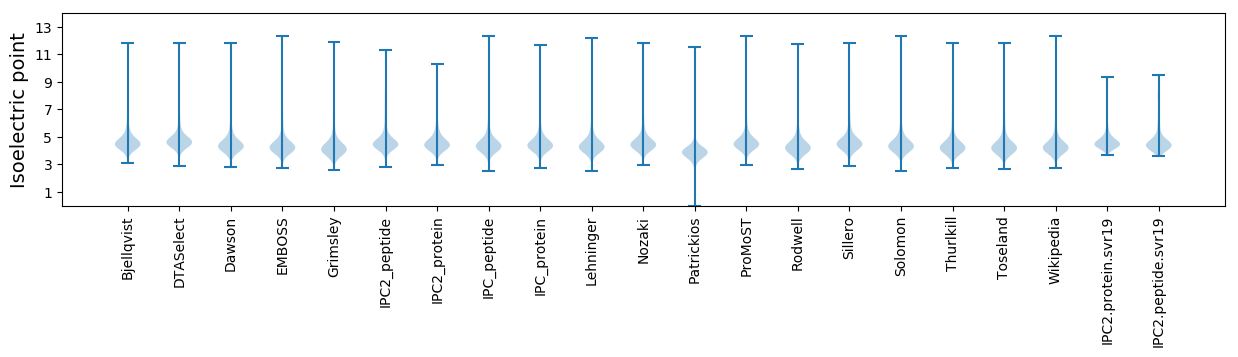
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q18KL4|Q18KL4_HALWD SnoaL-like domain-containing protein OS=Haloquadratum walsbyi (strain DSM 16790 / HBSQ001) OX=362976 GN=HQ_1303A PE=4 SV=1
MM1 pKa = 7.53ARR3 pKa = 11.84NEE5 pKa = 3.96AAIKK9 pKa = 9.23RR10 pKa = 11.84TLEE13 pKa = 3.63KK14 pKa = 10.14QICMRR19 pKa = 11.84CNARR23 pKa = 11.84NPQRR27 pKa = 11.84ASEE30 pKa = 4.11CRR32 pKa = 11.84KK33 pKa = 9.96CGYY36 pKa = 10.66SNLRR40 pKa = 11.84PKK42 pKa = 10.65SKK44 pKa = 10.08EE45 pKa = 3.37RR46 pKa = 11.84RR47 pKa = 11.84AAA49 pKa = 3.42
MM1 pKa = 7.53ARR3 pKa = 11.84NEE5 pKa = 3.96AAIKK9 pKa = 9.23RR10 pKa = 11.84TLEE13 pKa = 3.63KK14 pKa = 10.14QICMRR19 pKa = 11.84CNARR23 pKa = 11.84NPQRR27 pKa = 11.84ASEE30 pKa = 4.11CRR32 pKa = 11.84KK33 pKa = 9.96CGYY36 pKa = 10.66SNLRR40 pKa = 11.84PKK42 pKa = 10.65SKK44 pKa = 10.08EE45 pKa = 3.37RR46 pKa = 11.84RR47 pKa = 11.84AAA49 pKa = 3.42
Molecular weight: 5.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
772216 |
19 |
9159 |
301.9 |
32.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.398 ± 0.053 | 0.755 ± 0.018 |
7.819 ± 0.057 | 7.451 ± 0.08 |
3.201 ± 0.038 | 7.495 ± 0.077 |
2.092 ± 0.029 | 6.198 ± 0.046 |
2.335 ± 0.033 | 8.184 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.811 ± 0.024 | 3.443 ± 0.048 |
4.358 ± 0.035 | 3.429 ± 0.031 |
5.77 ± 0.049 | 6.82 ± 0.066 |
7.898 ± 0.08 | 7.801 ± 0.048 |
1.006 ± 0.019 | 2.736 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
