
Rhizobium meliloti (strain 1021) (Ensifer meliloti) (Sinorhizobium meliloti)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Rhizobiaceae; Sinorhizobium/Ensifer group; Sinorhizobium; Sinorhizobium meliloti
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
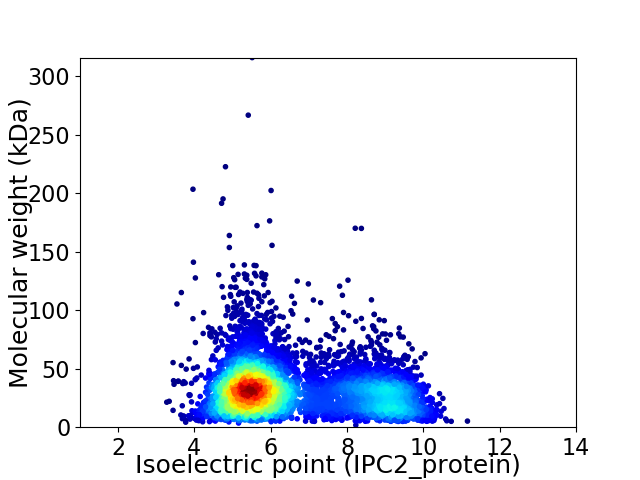
Virtual 2D-PAGE plot for 6169 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q92KC5|Q92KC5_RHIME Uncharacterized protein OS=Rhizobium meliloti (strain 1021) OX=266834 GN=SMc00940 PE=4 SV=1
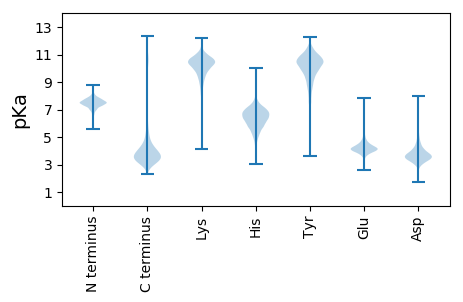
MM1 pKa = 7.62LIDD4 pKa = 4.54DD5 pKa = 4.05QPEE8 pKa = 3.66ISMVLPKK15 pKa = 10.27GRR17 pKa = 11.84AMATRR22 pKa = 11.84IYY24 pKa = 8.21GTNYY28 pKa = 10.59ADD30 pKa = 4.09IIKK33 pKa = 10.28QNGYY37 pKa = 9.43IAIEE41 pKa = 4.13IYY43 pKa = 10.42AYY45 pKa = 10.66DD46 pKa = 4.56GDD48 pKa = 4.58DD49 pKa = 5.02DD50 pKa = 4.92IYY52 pKa = 11.62LNRR55 pKa = 11.84TDD57 pKa = 4.95SYY59 pKa = 11.57GGYY62 pKa = 9.83NFVDD66 pKa = 3.04AGYY69 pKa = 11.4GNDD72 pKa = 3.56LVVNSYY78 pKa = 10.96EE79 pKa = 4.22GGNDD83 pKa = 2.84IYY85 pKa = 11.24LGGGNDD91 pKa = 3.63IYY93 pKa = 11.46VADD96 pKa = 3.61IRR98 pKa = 11.84VRR100 pKa = 11.84DD101 pKa = 3.7ASSYY105 pKa = 10.3DD106 pKa = 3.14IVYY109 pKa = 10.39GGSGNDD115 pKa = 2.85RR116 pKa = 11.84FEE118 pKa = 5.18IEE120 pKa = 5.36GYY122 pKa = 10.62ASDD125 pKa = 4.55YY126 pKa = 10.89YY127 pKa = 11.36GEE129 pKa = 4.58SGNDD133 pKa = 3.1TFFSVGFNNYY143 pKa = 9.05FNGGTGTDD151 pKa = 3.56TISYY155 pKa = 9.27QFQDD159 pKa = 2.75DD160 pKa = 3.38WSAEE164 pKa = 3.97RR165 pKa = 11.84GKK167 pKa = 11.1GVTIDD172 pKa = 3.85LGYY175 pKa = 11.18NYY177 pKa = 8.51ATTGSGRR184 pKa = 11.84RR185 pKa = 11.84EE186 pKa = 3.75DD187 pKa = 4.76LISIEE192 pKa = 4.21NATGTNYY199 pKa = 10.38GHH201 pKa = 7.85DD202 pKa = 5.48DD203 pKa = 3.3ITGSAVANTLRR214 pKa = 11.84GLGGHH219 pKa = 7.18DD220 pKa = 3.55IIEE223 pKa = 4.45GLGGNDD229 pKa = 3.39VLDD232 pKa = 4.69GGSGDD237 pKa = 4.09DD238 pKa = 4.32DD239 pKa = 5.55LYY241 pKa = 11.4GGSGADD247 pKa = 3.28ILRR250 pKa = 11.84GGTGFDD256 pKa = 3.64YY257 pKa = 11.14LVGGTGTDD265 pKa = 3.0SFDD268 pKa = 3.78FNSISEE274 pKa = 4.45SAVGSRR280 pKa = 11.84RR281 pKa = 11.84DD282 pKa = 3.6VITDD286 pKa = 3.89FHH288 pKa = 8.09RR289 pKa = 11.84SEE291 pKa = 4.63FDD293 pKa = 3.7VIDD296 pKa = 4.92LSTIDD301 pKa = 6.04ADD303 pKa = 4.42TTWSGNQSFTYY314 pKa = 10.01IGGNAFSGEE323 pKa = 3.92AGEE326 pKa = 4.54LNFRR330 pKa = 11.84SGIISGDD337 pKa = 3.64VNGDD341 pKa = 3.43GYY343 pKa = 11.78ADD345 pKa = 3.46FQVRR349 pKa = 11.84VNGATSLRR357 pKa = 11.84VDD359 pKa = 4.43DD360 pKa = 5.57FFLL363 pKa = 5.77
MM1 pKa = 7.62LIDD4 pKa = 4.54DD5 pKa = 4.05QPEE8 pKa = 3.66ISMVLPKK15 pKa = 10.27GRR17 pKa = 11.84AMATRR22 pKa = 11.84IYY24 pKa = 8.21GTNYY28 pKa = 10.59ADD30 pKa = 4.09IIKK33 pKa = 10.28QNGYY37 pKa = 9.43IAIEE41 pKa = 4.13IYY43 pKa = 10.42AYY45 pKa = 10.66DD46 pKa = 4.56GDD48 pKa = 4.58DD49 pKa = 5.02DD50 pKa = 4.92IYY52 pKa = 11.62LNRR55 pKa = 11.84TDD57 pKa = 4.95SYY59 pKa = 11.57GGYY62 pKa = 9.83NFVDD66 pKa = 3.04AGYY69 pKa = 11.4GNDD72 pKa = 3.56LVVNSYY78 pKa = 10.96EE79 pKa = 4.22GGNDD83 pKa = 2.84IYY85 pKa = 11.24LGGGNDD91 pKa = 3.63IYY93 pKa = 11.46VADD96 pKa = 3.61IRR98 pKa = 11.84VRR100 pKa = 11.84DD101 pKa = 3.7ASSYY105 pKa = 10.3DD106 pKa = 3.14IVYY109 pKa = 10.39GGSGNDD115 pKa = 2.85RR116 pKa = 11.84FEE118 pKa = 5.18IEE120 pKa = 5.36GYY122 pKa = 10.62ASDD125 pKa = 4.55YY126 pKa = 10.89YY127 pKa = 11.36GEE129 pKa = 4.58SGNDD133 pKa = 3.1TFFSVGFNNYY143 pKa = 9.05FNGGTGTDD151 pKa = 3.56TISYY155 pKa = 9.27QFQDD159 pKa = 2.75DD160 pKa = 3.38WSAEE164 pKa = 3.97RR165 pKa = 11.84GKK167 pKa = 11.1GVTIDD172 pKa = 3.85LGYY175 pKa = 11.18NYY177 pKa = 8.51ATTGSGRR184 pKa = 11.84RR185 pKa = 11.84EE186 pKa = 3.75DD187 pKa = 4.76LISIEE192 pKa = 4.21NATGTNYY199 pKa = 10.38GHH201 pKa = 7.85DD202 pKa = 5.48DD203 pKa = 3.3ITGSAVANTLRR214 pKa = 11.84GLGGHH219 pKa = 7.18DD220 pKa = 3.55IIEE223 pKa = 4.45GLGGNDD229 pKa = 3.39VLDD232 pKa = 4.69GGSGDD237 pKa = 4.09DD238 pKa = 4.32DD239 pKa = 5.55LYY241 pKa = 11.4GGSGADD247 pKa = 3.28ILRR250 pKa = 11.84GGTGFDD256 pKa = 3.64YY257 pKa = 11.14LVGGTGTDD265 pKa = 3.0SFDD268 pKa = 3.78FNSISEE274 pKa = 4.45SAVGSRR280 pKa = 11.84RR281 pKa = 11.84DD282 pKa = 3.6VITDD286 pKa = 3.89FHH288 pKa = 8.09RR289 pKa = 11.84SEE291 pKa = 4.63FDD293 pKa = 3.7VIDD296 pKa = 4.92LSTIDD301 pKa = 6.04ADD303 pKa = 4.42TTWSGNQSFTYY314 pKa = 10.01IGGNAFSGEE323 pKa = 3.92AGEE326 pKa = 4.54LNFRR330 pKa = 11.84SGIISGDD337 pKa = 3.64VNGDD341 pKa = 3.43GYY343 pKa = 11.78ADD345 pKa = 3.46FQVRR349 pKa = 11.84VNGATSLRR357 pKa = 11.84VDD359 pKa = 4.43DD360 pKa = 5.57FFLL363 pKa = 5.77
Molecular weight: 38.9 kDa
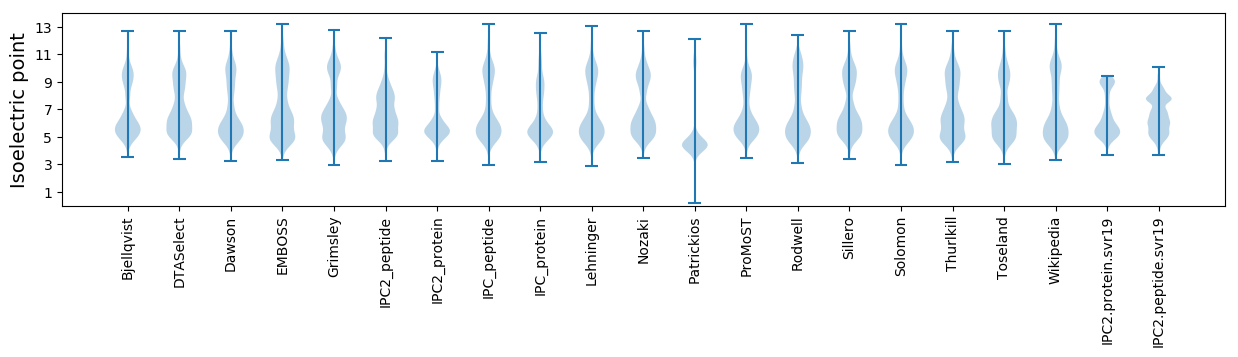
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q92SH8|Q92SH8_RHIME Ribokinase OS=Rhizobium meliloti (strain 1021) OX=266834 GN=rbsK PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.46GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 10.06GGRR28 pKa = 11.84KK29 pKa = 9.01VLTARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.46GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 10.06GGRR28 pKa = 11.84KK29 pKa = 9.01VLTARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
Molecular weight: 5.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1905276 |
14 |
2832 |
308.8 |
33.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.124 ± 0.035 | 0.861 ± 0.011 |
5.494 ± 0.023 | 6.037 ± 0.032 |
3.914 ± 0.022 | 8.538 ± 0.039 |
2.046 ± 0.016 | 5.477 ± 0.021 |
3.419 ± 0.023 | 10.021 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.519 ± 0.015 | 2.673 ± 0.017 |
4.958 ± 0.021 | 2.898 ± 0.016 |
7.169 ± 0.035 | 5.664 ± 0.02 |
5.157 ± 0.021 | 7.455 ± 0.024 |
1.299 ± 0.012 | 2.276 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
