
Mus musculus polyomavirus 3
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Betapolyomavirus
Average proteome isoelectric point is 7.34
Get precalculated fractions of proteins
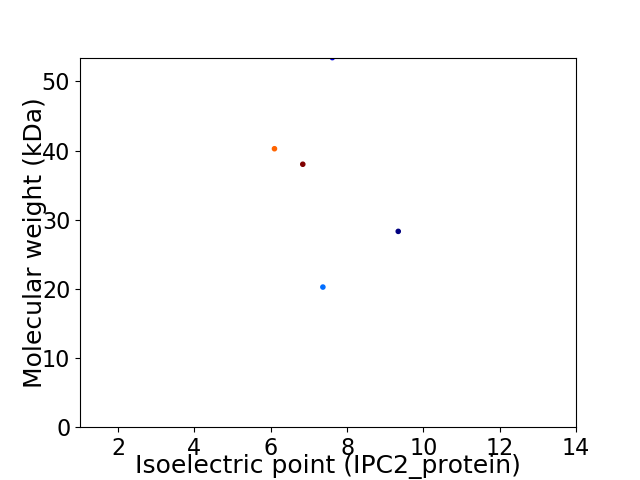
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
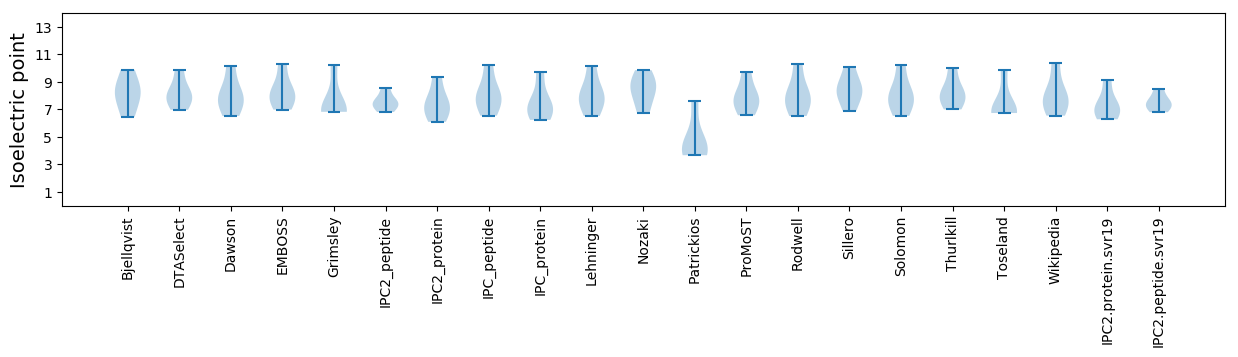
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2S0SYZ9|A0A2S0SYZ9_9POLY VP3 OS=Mus musculus polyomavirus 3 OX=2171394 PE=4 SV=1
MM1 pKa = 7.41GALLAIPEE9 pKa = 4.98LIAAAVEE16 pKa = 4.5GGAEE20 pKa = 4.45AIAIAGSEE28 pKa = 4.26SAILTGEE35 pKa = 4.1GAANLGLLSEE45 pKa = 4.56SAAVLDD51 pKa = 4.35GSIEE55 pKa = 3.9VSAAAEE61 pKa = 4.1SVLVNTPKK69 pKa = 10.36VVPEE73 pKa = 4.84LIAIQGGVQAGAGLTGGIASYY94 pKa = 10.75FLTADD99 pKa = 3.67SPGEE103 pKa = 3.97LPAQQHH109 pKa = 5.03TSGGRR114 pKa = 11.84DD115 pKa = 3.24VQGYY119 pKa = 9.83NGAQGSMALQGIPLHH134 pKa = 6.2ALEE137 pKa = 5.38HH138 pKa = 5.95GVPGIPDD145 pKa = 3.34WVLQLVPDD153 pKa = 4.79LPSLQDD159 pKa = 2.94IVYY162 pKa = 9.28RR163 pKa = 11.84IASGIVSSYY172 pKa = 7.83YY173 pKa = 8.39HH174 pKa = 6.2TGRR177 pKa = 11.84VIVQRR182 pKa = 11.84ALSEE186 pKa = 4.0EE187 pKa = 4.14MQRR190 pKa = 11.84LLNDD194 pKa = 3.43LSDD197 pKa = 3.5GFRR200 pKa = 11.84YY201 pKa = 8.35TFEE204 pKa = 3.97TLGRR208 pKa = 11.84SDD210 pKa = 4.16PVGAMIDD217 pKa = 3.87QIQSAHH223 pKa = 6.31RR224 pKa = 11.84YY225 pKa = 6.61FRR227 pKa = 11.84RR228 pKa = 11.84RR229 pKa = 11.84EE230 pKa = 3.71QMLLIEE236 pKa = 5.1GSDD239 pKa = 3.95PIPFGEE245 pKa = 4.02IASRR249 pKa = 11.84AGEE252 pKa = 3.82IAGNVAEE259 pKa = 4.44TVSNVVVDD267 pKa = 3.95VANLPRR273 pKa = 11.84DD274 pKa = 3.75GYY276 pKa = 10.71NALSEE281 pKa = 4.77GIHH284 pKa = 7.17RR285 pKa = 11.84LGQWVQYY292 pKa = 10.28AGSDD296 pKa = 3.47GGTGHH301 pKa = 6.91YY302 pKa = 7.58STPSWILYY310 pKa = 8.42VLQEE314 pKa = 4.73LEE316 pKa = 4.19RR317 pKa = 11.84EE318 pKa = 4.24LPKK321 pKa = 10.29IPRR324 pKa = 11.84TSLKK328 pKa = 10.7RR329 pKa = 11.84KK330 pKa = 8.91DD331 pKa = 3.4TDD333 pKa = 3.29NVHH336 pKa = 6.36NNNRR340 pKa = 11.84PKK342 pKa = 10.58KK343 pKa = 9.93ARR345 pKa = 11.84ARR347 pKa = 11.84KK348 pKa = 9.31NPSGPKK354 pKa = 9.67ASKK357 pKa = 10.4SNQKK361 pKa = 9.58RR362 pKa = 11.84RR363 pKa = 11.84SGSVRR368 pKa = 11.84RR369 pKa = 11.84RR370 pKa = 11.84STRR373 pKa = 11.84TNNRR377 pKa = 11.84VV378 pKa = 2.83
MM1 pKa = 7.41GALLAIPEE9 pKa = 4.98LIAAAVEE16 pKa = 4.5GGAEE20 pKa = 4.45AIAIAGSEE28 pKa = 4.26SAILTGEE35 pKa = 4.1GAANLGLLSEE45 pKa = 4.56SAAVLDD51 pKa = 4.35GSIEE55 pKa = 3.9VSAAAEE61 pKa = 4.1SVLVNTPKK69 pKa = 10.36VVPEE73 pKa = 4.84LIAIQGGVQAGAGLTGGIASYY94 pKa = 10.75FLTADD99 pKa = 3.67SPGEE103 pKa = 3.97LPAQQHH109 pKa = 5.03TSGGRR114 pKa = 11.84DD115 pKa = 3.24VQGYY119 pKa = 9.83NGAQGSMALQGIPLHH134 pKa = 6.2ALEE137 pKa = 5.38HH138 pKa = 5.95GVPGIPDD145 pKa = 3.34WVLQLVPDD153 pKa = 4.79LPSLQDD159 pKa = 2.94IVYY162 pKa = 9.28RR163 pKa = 11.84IASGIVSSYY172 pKa = 7.83YY173 pKa = 8.39HH174 pKa = 6.2TGRR177 pKa = 11.84VIVQRR182 pKa = 11.84ALSEE186 pKa = 4.0EE187 pKa = 4.14MQRR190 pKa = 11.84LLNDD194 pKa = 3.43LSDD197 pKa = 3.5GFRR200 pKa = 11.84YY201 pKa = 8.35TFEE204 pKa = 3.97TLGRR208 pKa = 11.84SDD210 pKa = 4.16PVGAMIDD217 pKa = 3.87QIQSAHH223 pKa = 6.31RR224 pKa = 11.84YY225 pKa = 6.61FRR227 pKa = 11.84RR228 pKa = 11.84RR229 pKa = 11.84EE230 pKa = 3.71QMLLIEE236 pKa = 5.1GSDD239 pKa = 3.95PIPFGEE245 pKa = 4.02IASRR249 pKa = 11.84AGEE252 pKa = 3.82IAGNVAEE259 pKa = 4.44TVSNVVVDD267 pKa = 3.95VANLPRR273 pKa = 11.84DD274 pKa = 3.75GYY276 pKa = 10.71NALSEE281 pKa = 4.77GIHH284 pKa = 7.17RR285 pKa = 11.84LGQWVQYY292 pKa = 10.28AGSDD296 pKa = 3.47GGTGHH301 pKa = 6.91YY302 pKa = 7.58STPSWILYY310 pKa = 8.42VLQEE314 pKa = 4.73LEE316 pKa = 4.19RR317 pKa = 11.84EE318 pKa = 4.24LPKK321 pKa = 10.29IPRR324 pKa = 11.84TSLKK328 pKa = 10.7RR329 pKa = 11.84KK330 pKa = 8.91DD331 pKa = 3.4TDD333 pKa = 3.29NVHH336 pKa = 6.36NNNRR340 pKa = 11.84PKK342 pKa = 10.58KK343 pKa = 9.93ARR345 pKa = 11.84ARR347 pKa = 11.84KK348 pKa = 9.31NPSGPKK354 pKa = 9.67ASKK357 pKa = 10.4SNQKK361 pKa = 9.58RR362 pKa = 11.84RR363 pKa = 11.84SGSVRR368 pKa = 11.84RR369 pKa = 11.84RR370 pKa = 11.84STRR373 pKa = 11.84TNNRR377 pKa = 11.84VV378 pKa = 2.83
Molecular weight: 40.27 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2S0SZ03|A0A2S0SZ03_9POLY Small t antigen OS=Mus musculus polyomavirus 3 OX=2171394 PE=4 SV=1
MM1 pKa = 7.71ALQGIPLHH9 pKa = 6.2ALEE12 pKa = 5.38HH13 pKa = 5.95GVPGIPDD20 pKa = 3.34WVLQLVPDD28 pKa = 4.79LPSLQDD34 pKa = 2.94IVYY37 pKa = 9.28RR38 pKa = 11.84IASGIVSSYY47 pKa = 7.83YY48 pKa = 8.39HH49 pKa = 6.2TGRR52 pKa = 11.84VIVQRR57 pKa = 11.84ALSEE61 pKa = 4.0EE62 pKa = 4.14MQRR65 pKa = 11.84LLNDD69 pKa = 3.43LSDD72 pKa = 3.5GFRR75 pKa = 11.84YY76 pKa = 8.35TFEE79 pKa = 3.97TLGRR83 pKa = 11.84SDD85 pKa = 4.16PVGAMIDD92 pKa = 3.87QIQSAHH98 pKa = 6.31RR99 pKa = 11.84YY100 pKa = 6.61FRR102 pKa = 11.84RR103 pKa = 11.84RR104 pKa = 11.84EE105 pKa = 3.71QMLLIEE111 pKa = 5.1GSDD114 pKa = 3.95PIPFGEE120 pKa = 4.02IASRR124 pKa = 11.84AGEE127 pKa = 3.82IAGNVAEE134 pKa = 4.44TVSNVVVDD142 pKa = 3.95VANLPRR148 pKa = 11.84DD149 pKa = 3.75GYY151 pKa = 10.71NALSEE156 pKa = 4.77GIHH159 pKa = 7.17RR160 pKa = 11.84LGQWVQYY167 pKa = 10.28AGSDD171 pKa = 3.47GGTGHH176 pKa = 6.91YY177 pKa = 7.58STPSWILYY185 pKa = 8.42VLQEE189 pKa = 4.73LEE191 pKa = 4.19RR192 pKa = 11.84EE193 pKa = 4.24LPKK196 pKa = 10.29IPRR199 pKa = 11.84TSLKK203 pKa = 10.7RR204 pKa = 11.84KK205 pKa = 8.91DD206 pKa = 3.4TDD208 pKa = 3.29NVHH211 pKa = 6.36NNNRR215 pKa = 11.84PKK217 pKa = 10.58KK218 pKa = 9.93ARR220 pKa = 11.84ARR222 pKa = 11.84KK223 pKa = 9.31NPSGPKK229 pKa = 9.67ASKK232 pKa = 10.4SNQKK236 pKa = 9.58RR237 pKa = 11.84RR238 pKa = 11.84SGSVRR243 pKa = 11.84RR244 pKa = 11.84RR245 pKa = 11.84STRR248 pKa = 11.84TNNRR252 pKa = 11.84VV253 pKa = 2.83
MM1 pKa = 7.71ALQGIPLHH9 pKa = 6.2ALEE12 pKa = 5.38HH13 pKa = 5.95GVPGIPDD20 pKa = 3.34WVLQLVPDD28 pKa = 4.79LPSLQDD34 pKa = 2.94IVYY37 pKa = 9.28RR38 pKa = 11.84IASGIVSSYY47 pKa = 7.83YY48 pKa = 8.39HH49 pKa = 6.2TGRR52 pKa = 11.84VIVQRR57 pKa = 11.84ALSEE61 pKa = 4.0EE62 pKa = 4.14MQRR65 pKa = 11.84LLNDD69 pKa = 3.43LSDD72 pKa = 3.5GFRR75 pKa = 11.84YY76 pKa = 8.35TFEE79 pKa = 3.97TLGRR83 pKa = 11.84SDD85 pKa = 4.16PVGAMIDD92 pKa = 3.87QIQSAHH98 pKa = 6.31RR99 pKa = 11.84YY100 pKa = 6.61FRR102 pKa = 11.84RR103 pKa = 11.84RR104 pKa = 11.84EE105 pKa = 3.71QMLLIEE111 pKa = 5.1GSDD114 pKa = 3.95PIPFGEE120 pKa = 4.02IASRR124 pKa = 11.84AGEE127 pKa = 3.82IAGNVAEE134 pKa = 4.44TVSNVVVDD142 pKa = 3.95VANLPRR148 pKa = 11.84DD149 pKa = 3.75GYY151 pKa = 10.71NALSEE156 pKa = 4.77GIHH159 pKa = 7.17RR160 pKa = 11.84LGQWVQYY167 pKa = 10.28AGSDD171 pKa = 3.47GGTGHH176 pKa = 6.91YY177 pKa = 7.58STPSWILYY185 pKa = 8.42VLQEE189 pKa = 4.73LEE191 pKa = 4.19RR192 pKa = 11.84EE193 pKa = 4.24LPKK196 pKa = 10.29IPRR199 pKa = 11.84TSLKK203 pKa = 10.7RR204 pKa = 11.84KK205 pKa = 8.91DD206 pKa = 3.4TDD208 pKa = 3.29NVHH211 pKa = 6.36NNNRR215 pKa = 11.84PKK217 pKa = 10.58KK218 pKa = 9.93ARR220 pKa = 11.84ARR222 pKa = 11.84KK223 pKa = 9.31NPSGPKK229 pKa = 9.67ASKK232 pKa = 10.4SNQKK236 pKa = 9.58RR237 pKa = 11.84RR238 pKa = 11.84SGSVRR243 pKa = 11.84RR244 pKa = 11.84RR245 pKa = 11.84STRR248 pKa = 11.84TNNRR252 pKa = 11.84VV253 pKa = 2.83
Molecular weight: 28.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1613 |
173 |
464 |
322.6 |
36.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.51 ± 1.016 | 2.48 ± 1.06 |
4.712 ± 0.502 | 5.952 ± 0.344 |
3.286 ± 0.788 | 7.75 ± 1.153 |
2.48 ± 0.324 | 5.022 ± 0.491 |
6.138 ± 1.661 | 9.361 ± 0.707 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.232 ± 0.288 | 5.084 ± 0.592 |
5.146 ± 0.769 | 4.154 ± 0.278 |
5.828 ± 1.086 | 6.324 ± 0.966 |
4.836 ± 0.771 | 7.688 ± 0.356 |
1.24 ± 0.316 | 3.782 ± 0.258 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
