
Cohaesibacter sp. ES.047
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Cohaesibacteraceae; Cohaesibacter; unclassified Cohaesibacter
Average proteome isoelectric point is 6.16
Get precalculated fractions of proteins
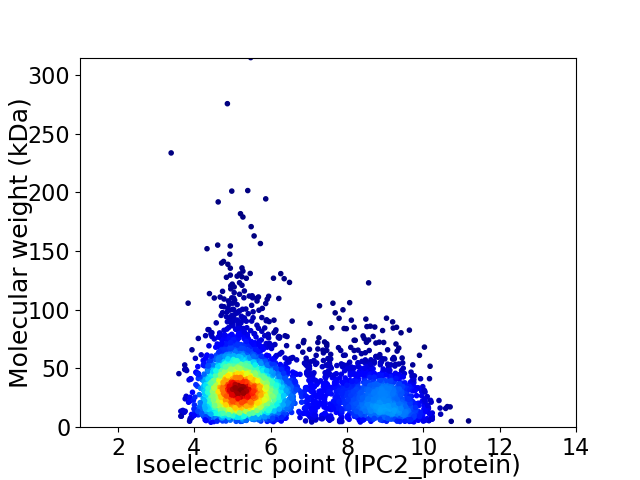
Virtual 2D-PAGE plot for 4413 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A285M074|A0A285M074_9RHIZ Histidine kinase OS=Cohaesibacter sp. ES.047 OX=1798205 GN=SAMN04515647_0760 PE=4 SV=1
MM1 pKa = 7.06ATTYY5 pKa = 11.19AVTEE9 pKa = 4.28SADD12 pKa = 3.75DD13 pKa = 3.92GTGNTSGTLSWAILQATSNGDD34 pKa = 3.58VIDD37 pKa = 3.91IDD39 pKa = 4.32ASVSSITITGSLPSASANISITTNHH64 pKa = 5.89AVEE67 pKa = 4.84VIGSALSGSGSTSLTLNGVSSAAALEE93 pKa = 4.28ATLSGTIQGHH103 pKa = 5.63NATSSGGSGSTALSGAGFILDD124 pKa = 3.54SSSNITGGKK133 pKa = 9.65ARR135 pKa = 11.84DD136 pKa = 3.69GASRR140 pKa = 11.84SYY142 pKa = 11.04SYY144 pKa = 11.58YY145 pKa = 10.66SGSNGQNGGAGGNGINAQAFSLEE168 pKa = 3.95SSGYY172 pKa = 9.38ISGGNGGNGGKK183 pKa = 9.82GASASYY189 pKa = 10.13SGRR192 pKa = 11.84AGGAGGAGGSAVSGYY207 pKa = 10.51SFSLEE212 pKa = 4.0TSGSVTGGSGGNGGNGGYY230 pKa = 10.24SLDD233 pKa = 3.57SSGGHH238 pKa = 6.0GGKK241 pKa = 10.2GAVGGASVSGTSFSLVTSGSVTGGSGGNGGAGGRR275 pKa = 11.84GGGRR279 pKa = 11.84NGGTGGIGGAGGTAISGTSFSLTNSGTLTGGDD311 pKa = 3.89AGDD314 pKa = 4.3GGNGGTSGSYY324 pKa = 10.61SSGLDD329 pKa = 3.26GGDD332 pKa = 3.16GSVGGTGGAGVSGSAFSLVNSGSITGGNGGAGGDD366 pKa = 3.85GGHH369 pKa = 6.52ATLNTTYY376 pKa = 10.66QIGGNGANGGSGGAGVSGDD395 pKa = 4.92AITLTNSGTITGGNGGAGGNAGGSGSGNAASDD427 pKa = 3.85GTDD430 pKa = 3.09GAGGVGVTGYY440 pKa = 10.71DD441 pKa = 2.95ISLVNSGTISGGLSAYY457 pKa = 10.49GLTRR461 pKa = 11.84ANAIEE466 pKa = 4.38FTSGTNSLKK475 pKa = 10.67LQSGYY480 pKa = 10.74VINGNVVANGSSDD493 pKa = 3.45TLILGGSDD501 pKa = 3.54DD502 pKa = 5.25ASFDD506 pKa = 3.99LDD508 pKa = 5.48DD509 pKa = 5.45IGSQYY514 pKa = 11.58AGFEE518 pKa = 4.54AYY520 pKa = 10.37SKK522 pKa = 9.46TGSSTWTLTGTAGEE536 pKa = 4.05AMGIFSISGGALAVNTVFDD555 pKa = 3.85SAISVLDD562 pKa = 3.66GGTLGGSGTVSNVTLYY578 pKa = 10.62SGSTIAPGNSIDD590 pKa = 3.86TLTVDD595 pKa = 3.12SDD597 pKa = 4.08LAFVSGSTYY606 pKa = 9.57EE607 pKa = 4.13VEE609 pKa = 5.12VDD611 pKa = 4.9DD612 pKa = 5.53EE613 pKa = 4.78GNSDD617 pKa = 3.8KK618 pKa = 11.02LVSTGTVSIEE628 pKa = 3.82EE629 pKa = 4.43GTTLSIIAEE638 pKa = 4.32NGTDD642 pKa = 3.79DD643 pKa = 3.9GSTYY647 pKa = 11.22AVNTDD652 pKa = 3.29YY653 pKa = 11.36SILSASSLSGAFSTIEE669 pKa = 3.66EE670 pKa = 4.2DD671 pKa = 4.52FAYY674 pKa = 10.99LDD676 pKa = 3.46ATVSYY681 pKa = 10.87NSDD684 pKa = 3.39EE685 pKa = 4.38AVLTLTRR692 pKa = 11.84ALGSSFADD700 pKa = 4.04FADD703 pKa = 3.52TTNQKK708 pKa = 9.98NVANVIEE715 pKa = 4.59GFGASDD721 pKa = 3.51SLYY724 pKa = 10.83DD725 pKa = 4.71AIVVLPDD732 pKa = 3.93GEE734 pKa = 4.34PVDD737 pKa = 5.35AFNQLTGEE745 pKa = 4.22QYY747 pKa = 10.08ATLQTIMTLNSAIDD761 pKa = 3.43RR762 pKa = 11.84NAANQRR768 pKa = 11.84LRR770 pKa = 11.84SSMGGIGGSGDD781 pKa = 3.48QVSVGFHH788 pKa = 6.06NEE790 pKa = 3.66EE791 pKa = 4.17EE792 pKa = 4.77LPSSLYY798 pKa = 7.63PTPQIWAQAFGNWSKK813 pKa = 10.78IGSTVNTASVTSQSKK828 pKa = 10.16GLLVGMDD835 pKa = 3.23AEE837 pKa = 4.58MWSDD841 pKa = 2.6WRR843 pKa = 11.84TGAFAGYY850 pKa = 9.39SQTSSKK856 pKa = 10.95ADD858 pKa = 3.95SINSAADD865 pKa = 3.0IDD867 pKa = 4.72HH868 pKa = 6.55IHH870 pKa = 6.8AGFYY874 pKa = 10.61AGTQWDD880 pKa = 3.87AFDD883 pKa = 5.59RR884 pKa = 11.84MVDD887 pKa = 3.7LNLGVSGVWHH897 pKa = 7.02HH898 pKa = 6.97IDD900 pKa = 3.68SEE902 pKa = 4.75RR903 pKa = 11.84DD904 pKa = 3.32VAFTGFSDD912 pKa = 5.21HH913 pKa = 6.64LTADD917 pKa = 3.59YY918 pKa = 11.15NAATTQAFAEE928 pKa = 3.91IGYY931 pKa = 8.41TYY933 pKa = 10.69EE934 pKa = 4.17LYY936 pKa = 10.31EE937 pKa = 4.73ARR939 pKa = 11.84LQPFLGAAIISQWSDD954 pKa = 2.96DD955 pKa = 3.59FTEE958 pKa = 4.23QGGAAALSAASATTVLGMTTLGARR982 pKa = 11.84GDD984 pKa = 3.73VQLGQFGGISASLTGSAAWQHH1005 pKa = 5.27VLGDD1009 pKa = 4.02VNSTTTMRR1017 pKa = 11.84FASGNDD1023 pKa = 3.01SFSIYY1028 pKa = 8.6GTPLDD1033 pKa = 4.0RR1034 pKa = 11.84DD1035 pKa = 3.58AAFLEE1040 pKa = 4.68AGVSFQQDD1048 pKa = 2.63EE1049 pKa = 4.41DD1050 pKa = 4.37FSLNVSYY1057 pKa = 10.86RR1058 pKa = 11.84GNFSEE1063 pKa = 4.47NANEE1067 pKa = 4.14QGLHH1071 pKa = 6.49AGFKK1075 pKa = 9.74YY1076 pKa = 10.43QFF1078 pKa = 3.62
MM1 pKa = 7.06ATTYY5 pKa = 11.19AVTEE9 pKa = 4.28SADD12 pKa = 3.75DD13 pKa = 3.92GTGNTSGTLSWAILQATSNGDD34 pKa = 3.58VIDD37 pKa = 3.91IDD39 pKa = 4.32ASVSSITITGSLPSASANISITTNHH64 pKa = 5.89AVEE67 pKa = 4.84VIGSALSGSGSTSLTLNGVSSAAALEE93 pKa = 4.28ATLSGTIQGHH103 pKa = 5.63NATSSGGSGSTALSGAGFILDD124 pKa = 3.54SSSNITGGKK133 pKa = 9.65ARR135 pKa = 11.84DD136 pKa = 3.69GASRR140 pKa = 11.84SYY142 pKa = 11.04SYY144 pKa = 11.58YY145 pKa = 10.66SGSNGQNGGAGGNGINAQAFSLEE168 pKa = 3.95SSGYY172 pKa = 9.38ISGGNGGNGGKK183 pKa = 9.82GASASYY189 pKa = 10.13SGRR192 pKa = 11.84AGGAGGAGGSAVSGYY207 pKa = 10.51SFSLEE212 pKa = 4.0TSGSVTGGSGGNGGNGGYY230 pKa = 10.24SLDD233 pKa = 3.57SSGGHH238 pKa = 6.0GGKK241 pKa = 10.2GAVGGASVSGTSFSLVTSGSVTGGSGGNGGAGGRR275 pKa = 11.84GGGRR279 pKa = 11.84NGGTGGIGGAGGTAISGTSFSLTNSGTLTGGDD311 pKa = 3.89AGDD314 pKa = 4.3GGNGGTSGSYY324 pKa = 10.61SSGLDD329 pKa = 3.26GGDD332 pKa = 3.16GSVGGTGGAGVSGSAFSLVNSGSITGGNGGAGGDD366 pKa = 3.85GGHH369 pKa = 6.52ATLNTTYY376 pKa = 10.66QIGGNGANGGSGGAGVSGDD395 pKa = 4.92AITLTNSGTITGGNGGAGGNAGGSGSGNAASDD427 pKa = 3.85GTDD430 pKa = 3.09GAGGVGVTGYY440 pKa = 10.71DD441 pKa = 2.95ISLVNSGTISGGLSAYY457 pKa = 10.49GLTRR461 pKa = 11.84ANAIEE466 pKa = 4.38FTSGTNSLKK475 pKa = 10.67LQSGYY480 pKa = 10.74VINGNVVANGSSDD493 pKa = 3.45TLILGGSDD501 pKa = 3.54DD502 pKa = 5.25ASFDD506 pKa = 3.99LDD508 pKa = 5.48DD509 pKa = 5.45IGSQYY514 pKa = 11.58AGFEE518 pKa = 4.54AYY520 pKa = 10.37SKK522 pKa = 9.46TGSSTWTLTGTAGEE536 pKa = 4.05AMGIFSISGGALAVNTVFDD555 pKa = 3.85SAISVLDD562 pKa = 3.66GGTLGGSGTVSNVTLYY578 pKa = 10.62SGSTIAPGNSIDD590 pKa = 3.86TLTVDD595 pKa = 3.12SDD597 pKa = 4.08LAFVSGSTYY606 pKa = 9.57EE607 pKa = 4.13VEE609 pKa = 5.12VDD611 pKa = 4.9DD612 pKa = 5.53EE613 pKa = 4.78GNSDD617 pKa = 3.8KK618 pKa = 11.02LVSTGTVSIEE628 pKa = 3.82EE629 pKa = 4.43GTTLSIIAEE638 pKa = 4.32NGTDD642 pKa = 3.79DD643 pKa = 3.9GSTYY647 pKa = 11.22AVNTDD652 pKa = 3.29YY653 pKa = 11.36SILSASSLSGAFSTIEE669 pKa = 3.66EE670 pKa = 4.2DD671 pKa = 4.52FAYY674 pKa = 10.99LDD676 pKa = 3.46ATVSYY681 pKa = 10.87NSDD684 pKa = 3.39EE685 pKa = 4.38AVLTLTRR692 pKa = 11.84ALGSSFADD700 pKa = 4.04FADD703 pKa = 3.52TTNQKK708 pKa = 9.98NVANVIEE715 pKa = 4.59GFGASDD721 pKa = 3.51SLYY724 pKa = 10.83DD725 pKa = 4.71AIVVLPDD732 pKa = 3.93GEE734 pKa = 4.34PVDD737 pKa = 5.35AFNQLTGEE745 pKa = 4.22QYY747 pKa = 10.08ATLQTIMTLNSAIDD761 pKa = 3.43RR762 pKa = 11.84NAANQRR768 pKa = 11.84LRR770 pKa = 11.84SSMGGIGGSGDD781 pKa = 3.48QVSVGFHH788 pKa = 6.06NEE790 pKa = 3.66EE791 pKa = 4.17EE792 pKa = 4.77LPSSLYY798 pKa = 7.63PTPQIWAQAFGNWSKK813 pKa = 10.78IGSTVNTASVTSQSKK828 pKa = 10.16GLLVGMDD835 pKa = 3.23AEE837 pKa = 4.58MWSDD841 pKa = 2.6WRR843 pKa = 11.84TGAFAGYY850 pKa = 9.39SQTSSKK856 pKa = 10.95ADD858 pKa = 3.95SINSAADD865 pKa = 3.0IDD867 pKa = 4.72HH868 pKa = 6.55IHH870 pKa = 6.8AGFYY874 pKa = 10.61AGTQWDD880 pKa = 3.87AFDD883 pKa = 5.59RR884 pKa = 11.84MVDD887 pKa = 3.7LNLGVSGVWHH897 pKa = 7.02HH898 pKa = 6.97IDD900 pKa = 3.68SEE902 pKa = 4.75RR903 pKa = 11.84DD904 pKa = 3.32VAFTGFSDD912 pKa = 5.21HH913 pKa = 6.64LTADD917 pKa = 3.59YY918 pKa = 11.15NAATTQAFAEE928 pKa = 3.91IGYY931 pKa = 8.41TYY933 pKa = 10.69EE934 pKa = 4.17LYY936 pKa = 10.31EE937 pKa = 4.73ARR939 pKa = 11.84LQPFLGAAIISQWSDD954 pKa = 2.96DD955 pKa = 3.59FTEE958 pKa = 4.23QGGAAALSAASATTVLGMTTLGARR982 pKa = 11.84GDD984 pKa = 3.73VQLGQFGGISASLTGSAAWQHH1005 pKa = 5.27VLGDD1009 pKa = 4.02VNSTTTMRR1017 pKa = 11.84FASGNDD1023 pKa = 3.01SFSIYY1028 pKa = 8.6GTPLDD1033 pKa = 4.0RR1034 pKa = 11.84DD1035 pKa = 3.58AAFLEE1040 pKa = 4.68AGVSFQQDD1048 pKa = 2.63EE1049 pKa = 4.41DD1050 pKa = 4.37FSLNVSYY1057 pKa = 10.86RR1058 pKa = 11.84GNFSEE1063 pKa = 4.47NANEE1067 pKa = 4.14QGLHH1071 pKa = 6.49AGFKK1075 pKa = 9.74YY1076 pKa = 10.43QFF1078 pKa = 3.62
Molecular weight: 105.6 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A285M2E1|A0A285M2E1_9RHIZ Multidrug resistance efflux pump OS=Cohaesibacter sp. ES.047 OX=1798205 GN=SAMN04515647_1553 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.2QPSRR9 pKa = 11.84LVRR12 pKa = 11.84KK13 pKa = 9.21RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.48GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.31NGRR28 pKa = 11.84QILARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.95GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.2QPSRR9 pKa = 11.84LVRR12 pKa = 11.84KK13 pKa = 9.21RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.48GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.31NGRR28 pKa = 11.84QILARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.95GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1404519 |
39 |
2833 |
318.3 |
34.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.5 ± 0.043 | 0.952 ± 0.012 |
6.069 ± 0.031 | 6.164 ± 0.034 |
3.911 ± 0.026 | 7.884 ± 0.03 |
2.114 ± 0.019 | 5.96 ± 0.029 |
4.373 ± 0.03 | 10.085 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.816 ± 0.018 | 3.137 ± 0.022 |
4.59 ± 0.026 | 3.383 ± 0.021 |
5.888 ± 0.03 | 6.267 ± 0.028 |
5.307 ± 0.026 | 6.974 ± 0.029 |
1.26 ± 0.015 | 2.366 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
