
Canine papillomavirus 22
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Taupapillomavirus; unclassified Taupapillomavirus
Average proteome isoelectric point is 6.25
Get precalculated fractions of proteins
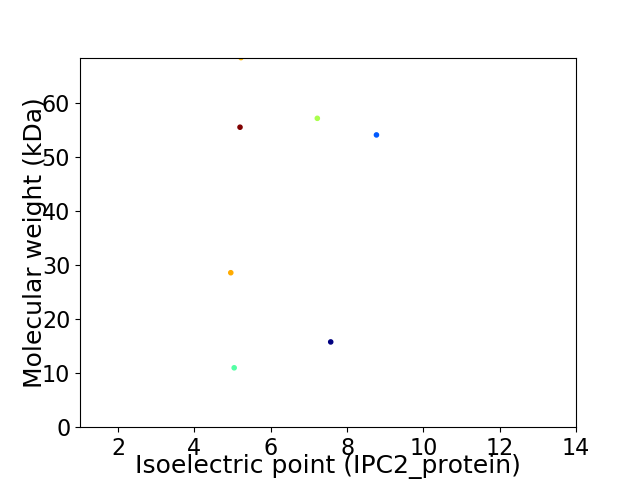
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346M198|A0A346M198_9PAPI Protein E6 OS=Canine papillomavirus 22 OX=2304620 GN=E6 PE=3 SV=1
MM1 pKa = 7.0MACIMRR7 pKa = 11.84TRR9 pKa = 11.84MEE11 pKa = 4.06NRR13 pKa = 11.84CIFAILKK20 pKa = 6.02QTPVALAPEE29 pKa = 4.91AYY31 pKa = 10.22GEE33 pKa = 4.08FCIRR37 pKa = 11.84MSACLPLFLARR48 pKa = 11.84DD49 pKa = 3.79PVPRR53 pKa = 11.84PPAAPTPGPGGDD65 pKa = 3.3RR66 pKa = 11.84IGPRR70 pKa = 11.84PHH72 pKa = 6.82NPQDD76 pKa = 3.82PNQAPPPQPDD86 pKa = 3.18TRR88 pKa = 11.84GDD90 pKa = 3.55RR91 pKa = 11.84QSPGGGEE98 pKa = 3.76QGPGDD103 pKa = 4.35EE104 pKa = 4.59EE105 pKa = 6.37DD106 pKa = 3.64EE107 pKa = 6.04DD108 pKa = 4.23EE109 pKa = 6.57DD110 pKa = 4.3EE111 pKa = 6.27DD112 pKa = 4.58EE113 pKa = 5.44DD114 pKa = 5.11RR115 pKa = 11.84PPPPPPPPAACNCQGRR131 pKa = 11.84TPQRR135 pKa = 11.84EE136 pKa = 3.47RR137 pKa = 11.84DD138 pKa = 3.06RR139 pKa = 11.84RR140 pKa = 11.84RR141 pKa = 11.84RR142 pKa = 11.84RR143 pKa = 11.84RR144 pKa = 11.84GRR146 pKa = 11.84ARR148 pKa = 11.84LRR150 pKa = 11.84FQGPEE155 pKa = 3.64GGEE158 pKa = 3.96EE159 pKa = 3.92EE160 pKa = 4.22KK161 pKa = 10.98AHH163 pKa = 6.07YY164 pKa = 10.08EE165 pKa = 3.89EE166 pKa = 6.43AGEE169 pKa = 4.19KK170 pKa = 10.79DD171 pKa = 3.55PDD173 pKa = 3.96EE174 pKa = 4.97EE175 pKa = 5.84DD176 pKa = 3.03PHH178 pKa = 7.26QPPTEE183 pKa = 4.07EE184 pKa = 4.28EE185 pKa = 4.48KK186 pKa = 11.14EE187 pKa = 3.83NSPPHH192 pKa = 6.39KK193 pKa = 9.38PAPLRR198 pKa = 11.84RR199 pKa = 11.84PYY201 pKa = 10.55YY202 pKa = 10.15SPPPGPLGDD211 pKa = 3.77STEE214 pKa = 4.97DD215 pKa = 3.79DD216 pKa = 4.21DD217 pKa = 5.67EE218 pKa = 6.8GPASLLKK225 pKa = 10.85DD226 pKa = 4.23LGRR229 pKa = 11.84KK230 pKa = 8.27WGADD234 pKa = 3.39LDD236 pKa = 4.22ALHH239 pKa = 6.54HH240 pKa = 6.4KK241 pKa = 9.85IWGDD245 pKa = 3.51LEE247 pKa = 4.08KK248 pKa = 11.14LRR250 pKa = 11.84DD251 pKa = 3.74RR252 pKa = 11.84LGASQQ257 pKa = 3.78
MM1 pKa = 7.0MACIMRR7 pKa = 11.84TRR9 pKa = 11.84MEE11 pKa = 4.06NRR13 pKa = 11.84CIFAILKK20 pKa = 6.02QTPVALAPEE29 pKa = 4.91AYY31 pKa = 10.22GEE33 pKa = 4.08FCIRR37 pKa = 11.84MSACLPLFLARR48 pKa = 11.84DD49 pKa = 3.79PVPRR53 pKa = 11.84PPAAPTPGPGGDD65 pKa = 3.3RR66 pKa = 11.84IGPRR70 pKa = 11.84PHH72 pKa = 6.82NPQDD76 pKa = 3.82PNQAPPPQPDD86 pKa = 3.18TRR88 pKa = 11.84GDD90 pKa = 3.55RR91 pKa = 11.84QSPGGGEE98 pKa = 3.76QGPGDD103 pKa = 4.35EE104 pKa = 4.59EE105 pKa = 6.37DD106 pKa = 3.64EE107 pKa = 6.04DD108 pKa = 4.23EE109 pKa = 6.57DD110 pKa = 4.3EE111 pKa = 6.27DD112 pKa = 4.58EE113 pKa = 5.44DD114 pKa = 5.11RR115 pKa = 11.84PPPPPPPPAACNCQGRR131 pKa = 11.84TPQRR135 pKa = 11.84EE136 pKa = 3.47RR137 pKa = 11.84DD138 pKa = 3.06RR139 pKa = 11.84RR140 pKa = 11.84RR141 pKa = 11.84RR142 pKa = 11.84RR143 pKa = 11.84RR144 pKa = 11.84GRR146 pKa = 11.84ARR148 pKa = 11.84LRR150 pKa = 11.84FQGPEE155 pKa = 3.64GGEE158 pKa = 3.96EE159 pKa = 3.92EE160 pKa = 4.22KK161 pKa = 10.98AHH163 pKa = 6.07YY164 pKa = 10.08EE165 pKa = 3.89EE166 pKa = 6.43AGEE169 pKa = 4.19KK170 pKa = 10.79DD171 pKa = 3.55PDD173 pKa = 3.96EE174 pKa = 4.97EE175 pKa = 5.84DD176 pKa = 3.03PHH178 pKa = 7.26QPPTEE183 pKa = 4.07EE184 pKa = 4.28EE185 pKa = 4.48KK186 pKa = 11.14EE187 pKa = 3.83NSPPHH192 pKa = 6.39KK193 pKa = 9.38PAPLRR198 pKa = 11.84RR199 pKa = 11.84PYY201 pKa = 10.55YY202 pKa = 10.15SPPPGPLGDD211 pKa = 3.77STEE214 pKa = 4.97DD215 pKa = 3.79DD216 pKa = 4.21DD217 pKa = 5.67EE218 pKa = 6.8GPASLLKK225 pKa = 10.85DD226 pKa = 4.23LGRR229 pKa = 11.84KK230 pKa = 8.27WGADD234 pKa = 3.39LDD236 pKa = 4.22ALHH239 pKa = 6.54HH240 pKa = 6.4KK241 pKa = 9.85IWGDD245 pKa = 3.51LEE247 pKa = 4.08KK248 pKa = 11.14LRR250 pKa = 11.84DD251 pKa = 3.74RR252 pKa = 11.84LGASQQ257 pKa = 3.78
Molecular weight: 28.55 kDa
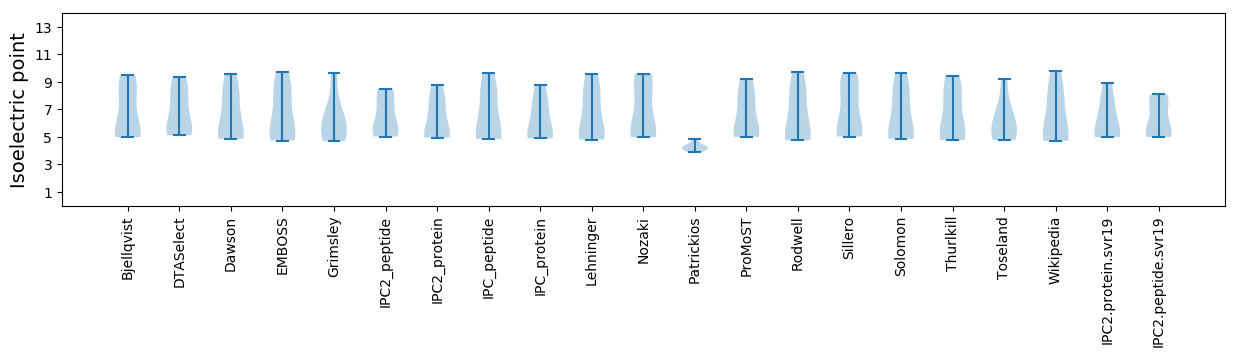
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346M197|A0A346M197_9PAPI E4 OS=Canine papillomavirus 22 OX=2304620 PE=4 SV=1
MM1 pKa = 6.81QNLRR5 pKa = 11.84IRR7 pKa = 11.84YY8 pKa = 8.63DD9 pKa = 3.41AVQEE13 pKa = 3.99ALFAHH18 pKa = 6.39YY19 pKa = 9.71EE20 pKa = 4.04QGSKK24 pKa = 10.54SLNDD28 pKa = 3.23QVSYY32 pKa = 10.32WEE34 pKa = 3.79LRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84EE39 pKa = 4.01SAILYY44 pKa = 6.54WARR47 pKa = 11.84KK48 pKa = 9.28NGISTLGFQPVPSLQSSEE66 pKa = 5.1ANAKK70 pKa = 9.68SAIGMSLTLQSLLASPFGQEE90 pKa = 3.56PWTLPEE96 pKa = 4.29TSLEE100 pKa = 4.1MWNTPPKK107 pKa = 10.91NCMKK111 pKa = 10.55KK112 pKa = 9.64HH113 pKa = 5.05GFTVEE118 pKa = 3.66VHH120 pKa = 6.56FDD122 pKa = 3.65NDD124 pKa = 3.66PEE126 pKa = 4.22NAFPYY131 pKa = 9.6TCWQSIYY138 pKa = 10.67YY139 pKa = 9.74QDD141 pKa = 4.39AEE143 pKa = 4.23TNEE146 pKa = 4.15WVKK149 pKa = 10.37TSGKK153 pKa = 9.1VAHH156 pKa = 7.18DD157 pKa = 3.21GMYY160 pKa = 10.87YY161 pKa = 10.05EE162 pKa = 5.78DD163 pKa = 4.93KK164 pKa = 11.01DD165 pKa = 3.8GEE167 pKa = 4.41QVYY170 pKa = 9.47FCHH173 pKa = 7.11FEE175 pKa = 3.86TDD177 pKa = 2.82ASRR180 pKa = 11.84FSTRR184 pKa = 11.84GIWRR188 pKa = 11.84VLYY191 pKa = 10.96KK192 pKa = 10.37NVCLSASVSSSGSGAPAAGGSNTGPWWGQDD222 pKa = 2.82WPEE225 pKa = 4.13APQPAGPQPSAASSTRR241 pKa = 11.84HH242 pKa = 5.25SRR244 pKa = 11.84GSPEE248 pKa = 3.53PRR250 pKa = 11.84RR251 pKa = 11.84RR252 pKa = 11.84GTGSRR257 pKa = 11.84GRR259 pKa = 11.84GGRR262 pKa = 11.84GRR264 pKa = 11.84GRR266 pKa = 11.84GRR268 pKa = 11.84GSSASPSAPACRR280 pKa = 11.84LQLPGPDD287 pKa = 3.48TPEE290 pKa = 5.62GEE292 pKa = 4.2GSEE295 pKa = 4.21EE296 pKa = 4.16EE297 pKa = 4.28EE298 pKa = 4.3EE299 pKa = 5.18GPCEE303 pKa = 4.15AEE305 pKa = 4.33VPRR308 pKa = 11.84SRR310 pKa = 11.84GRR312 pKa = 11.84GGGKK316 pKa = 9.94GPLRR320 pKa = 11.84GGRR323 pKa = 11.84GKK325 pKa = 10.4RR326 pKa = 11.84PRR328 pKa = 11.84RR329 pKa = 11.84GGPSPTTYY337 pKa = 10.05RR338 pKa = 11.84GRR340 pKa = 11.84EE341 pKa = 4.34GEE343 pKa = 4.0QSTSQTCTPPAPILQPSTGSTGGLYY368 pKa = 10.04RR369 pKa = 11.84GRR371 pKa = 11.84RR372 pKa = 11.84RR373 pKa = 11.84GPCLSPEE380 pKa = 4.34GSWSKK385 pKa = 10.66VGCRR389 pKa = 11.84PRR391 pKa = 11.84RR392 pKa = 11.84SSSQDD397 pKa = 2.96LGGPGEE403 pKa = 4.51ASGQTGRR410 pKa = 11.84VPIVVVKK417 pKa = 9.29GEE419 pKa = 4.21CNALKK424 pKa = 10.29CYY426 pKa = 10.24RR427 pKa = 11.84LRR429 pKa = 11.84LRR431 pKa = 11.84RR432 pKa = 11.84RR433 pKa = 11.84HH434 pKa = 5.26RR435 pKa = 11.84HH436 pKa = 4.03TFDD439 pKa = 4.91FISTGFGWIGGEE451 pKa = 3.86GRR453 pKa = 11.84KK454 pKa = 9.59VGRR457 pKa = 11.84QRR459 pKa = 11.84MLIAFSGYY467 pKa = 7.48DD468 pKa = 3.09QRR470 pKa = 11.84DD471 pKa = 3.44FFLGHH476 pKa = 5.32VKK478 pKa = 9.88IPKK481 pKa = 10.32LMDD484 pKa = 3.23VSLGEE489 pKa = 4.25LDD491 pKa = 3.81SLL493 pKa = 4.86
MM1 pKa = 6.81QNLRR5 pKa = 11.84IRR7 pKa = 11.84YY8 pKa = 8.63DD9 pKa = 3.41AVQEE13 pKa = 3.99ALFAHH18 pKa = 6.39YY19 pKa = 9.71EE20 pKa = 4.04QGSKK24 pKa = 10.54SLNDD28 pKa = 3.23QVSYY32 pKa = 10.32WEE34 pKa = 3.79LRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84EE39 pKa = 4.01SAILYY44 pKa = 6.54WARR47 pKa = 11.84KK48 pKa = 9.28NGISTLGFQPVPSLQSSEE66 pKa = 5.1ANAKK70 pKa = 9.68SAIGMSLTLQSLLASPFGQEE90 pKa = 3.56PWTLPEE96 pKa = 4.29TSLEE100 pKa = 4.1MWNTPPKK107 pKa = 10.91NCMKK111 pKa = 10.55KK112 pKa = 9.64HH113 pKa = 5.05GFTVEE118 pKa = 3.66VHH120 pKa = 6.56FDD122 pKa = 3.65NDD124 pKa = 3.66PEE126 pKa = 4.22NAFPYY131 pKa = 9.6TCWQSIYY138 pKa = 10.67YY139 pKa = 9.74QDD141 pKa = 4.39AEE143 pKa = 4.23TNEE146 pKa = 4.15WVKK149 pKa = 10.37TSGKK153 pKa = 9.1VAHH156 pKa = 7.18DD157 pKa = 3.21GMYY160 pKa = 10.87YY161 pKa = 10.05EE162 pKa = 5.78DD163 pKa = 4.93KK164 pKa = 11.01DD165 pKa = 3.8GEE167 pKa = 4.41QVYY170 pKa = 9.47FCHH173 pKa = 7.11FEE175 pKa = 3.86TDD177 pKa = 2.82ASRR180 pKa = 11.84FSTRR184 pKa = 11.84GIWRR188 pKa = 11.84VLYY191 pKa = 10.96KK192 pKa = 10.37NVCLSASVSSSGSGAPAAGGSNTGPWWGQDD222 pKa = 2.82WPEE225 pKa = 4.13APQPAGPQPSAASSTRR241 pKa = 11.84HH242 pKa = 5.25SRR244 pKa = 11.84GSPEE248 pKa = 3.53PRR250 pKa = 11.84RR251 pKa = 11.84RR252 pKa = 11.84GTGSRR257 pKa = 11.84GRR259 pKa = 11.84GGRR262 pKa = 11.84GRR264 pKa = 11.84GRR266 pKa = 11.84GRR268 pKa = 11.84GSSASPSAPACRR280 pKa = 11.84LQLPGPDD287 pKa = 3.48TPEE290 pKa = 5.62GEE292 pKa = 4.2GSEE295 pKa = 4.21EE296 pKa = 4.16EE297 pKa = 4.28EE298 pKa = 4.3EE299 pKa = 5.18GPCEE303 pKa = 4.15AEE305 pKa = 4.33VPRR308 pKa = 11.84SRR310 pKa = 11.84GRR312 pKa = 11.84GGGKK316 pKa = 9.94GPLRR320 pKa = 11.84GGRR323 pKa = 11.84GKK325 pKa = 10.4RR326 pKa = 11.84PRR328 pKa = 11.84RR329 pKa = 11.84GGPSPTTYY337 pKa = 10.05RR338 pKa = 11.84GRR340 pKa = 11.84EE341 pKa = 4.34GEE343 pKa = 4.0QSTSQTCTPPAPILQPSTGSTGGLYY368 pKa = 10.04RR369 pKa = 11.84GRR371 pKa = 11.84RR372 pKa = 11.84RR373 pKa = 11.84GPCLSPEE380 pKa = 4.34GSWSKK385 pKa = 10.66VGCRR389 pKa = 11.84PRR391 pKa = 11.84RR392 pKa = 11.84SSSQDD397 pKa = 2.96LGGPGEE403 pKa = 4.51ASGQTGRR410 pKa = 11.84VPIVVVKK417 pKa = 9.29GEE419 pKa = 4.21CNALKK424 pKa = 10.29CYY426 pKa = 10.24RR427 pKa = 11.84LRR429 pKa = 11.84LRR431 pKa = 11.84RR432 pKa = 11.84RR433 pKa = 11.84HH434 pKa = 5.26RR435 pKa = 11.84HH436 pKa = 4.03TFDD439 pKa = 4.91FISTGFGWIGGEE451 pKa = 3.86GRR453 pKa = 11.84KK454 pKa = 9.59VGRR457 pKa = 11.84QRR459 pKa = 11.84MLIAFSGYY467 pKa = 7.48DD468 pKa = 3.09QRR470 pKa = 11.84DD471 pKa = 3.44FFLGHH476 pKa = 5.32VKK478 pKa = 9.88IPKK481 pKa = 10.32LMDD484 pKa = 3.23VSLGEE489 pKa = 4.25LDD491 pKa = 3.81SLL493 pKa = 4.86
Molecular weight: 54.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2616 |
96 |
605 |
373.7 |
41.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.422 ± 0.567 | 2.599 ± 0.81 |
6.537 ± 0.788 | 6.307 ± 0.73 |
4.243 ± 0.764 | 8.142 ± 1.311 |
2.561 ± 0.38 | 3.593 ± 0.63 |
4.778 ± 0.86 | 8.142 ± 1.077 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.606 ± 0.433 | 3.211 ± 0.742 |
7.913 ± 1.438 | 4.434 ± 0.442 |
6.804 ± 1.075 | 7.645 ± 1.016 |
5.696 ± 0.453 | 5.657 ± 0.919 |
1.376 ± 0.311 | 2.332 ± 0.411 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
