
Streptomyces phyllanthi
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
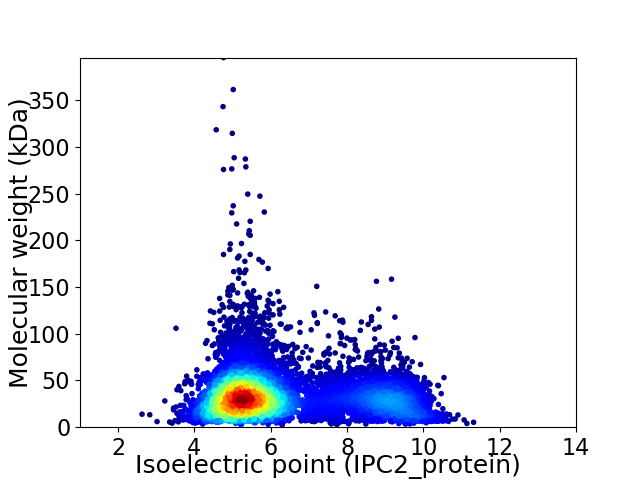
Virtual 2D-PAGE plot for 8637 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5N8W7H7|A0A5N8W7H7_9ACTN Sugar porter family MFS transporter OS=Streptomyces phyllanthi OX=1803180 GN=FNH04_27025 PE=3 SV=1
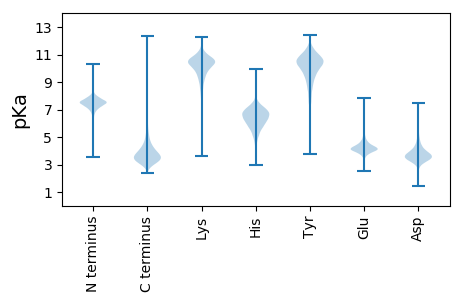
MM1 pKa = 6.89TQPPGQPPQDD11 pKa = 3.66GYY13 pKa = 11.59GAQPPQQPPYY23 pKa = 8.79PGPYY27 pKa = 9.45SPPPQPPTQVPGQSAQPGQPGQPPNPYY54 pKa = 9.97AAPQPQGPYY63 pKa = 10.05GAPQQPYY70 pKa = 9.8GAPQPPYY77 pKa = 10.16AGAPGAPGTPGMPGMPNYY95 pKa = 8.83GAPYY99 pKa = 9.64GGYY102 pKa = 9.26PPPPPPGGGGSKK114 pKa = 10.49NRR116 pKa = 11.84MALIIVGAVTAVAVVVGGIAVATSGSDD143 pKa = 3.3EE144 pKa = 4.44SSGATSDD151 pKa = 3.84KK152 pKa = 10.87NVSAEE157 pKa = 3.86PSVSDD162 pKa = 3.94GASEE166 pKa = 3.99EE167 pKa = 4.37AADD170 pKa = 4.9DD171 pKa = 4.37PSTEE175 pKa = 4.11ATDD178 pKa = 4.23DD179 pKa = 4.33PSDD182 pKa = 3.55SGYY185 pKa = 11.2SDD187 pKa = 3.55DD188 pKa = 4.02TGGGSEE194 pKa = 4.38EE195 pKa = 4.48APPEE199 pKa = 4.04PSVEE203 pKa = 4.29GQWQDD208 pKa = 3.55ADD210 pKa = 3.86AKK212 pKa = 9.52TLTVGTKK219 pKa = 8.33YY220 pKa = 9.53TSGDD224 pKa = 3.44LKK226 pKa = 11.03GKK228 pKa = 10.0YY229 pKa = 9.29ALSYY233 pKa = 10.72IDD235 pKa = 3.55TTGKK239 pKa = 10.87GILTGLGAYY248 pKa = 10.0RR249 pKa = 11.84DD250 pKa = 4.01DD251 pKa = 4.41DD252 pKa = 3.92SFRR255 pKa = 11.84LVLRR259 pKa = 11.84PMDD262 pKa = 4.45SDD264 pKa = 4.11SKK266 pKa = 11.55DD267 pKa = 3.4EE268 pKa = 3.99SDD270 pKa = 3.73YY271 pKa = 11.57VYY273 pKa = 10.29GTVRR277 pKa = 11.84RR278 pKa = 11.84SGDD281 pKa = 3.43DD282 pKa = 4.18VVITWDD288 pKa = 3.75DD289 pKa = 3.55GGKK292 pKa = 8.15EE293 pKa = 3.87TLGYY297 pKa = 10.05IGSLSDD303 pKa = 3.22
MM1 pKa = 6.89TQPPGQPPQDD11 pKa = 3.66GYY13 pKa = 11.59GAQPPQQPPYY23 pKa = 8.79PGPYY27 pKa = 9.45SPPPQPPTQVPGQSAQPGQPGQPPNPYY54 pKa = 9.97AAPQPQGPYY63 pKa = 10.05GAPQQPYY70 pKa = 9.8GAPQPPYY77 pKa = 10.16AGAPGAPGTPGMPGMPNYY95 pKa = 8.83GAPYY99 pKa = 9.64GGYY102 pKa = 9.26PPPPPPGGGGSKK114 pKa = 10.49NRR116 pKa = 11.84MALIIVGAVTAVAVVVGGIAVATSGSDD143 pKa = 3.3EE144 pKa = 4.44SSGATSDD151 pKa = 3.84KK152 pKa = 10.87NVSAEE157 pKa = 3.86PSVSDD162 pKa = 3.94GASEE166 pKa = 3.99EE167 pKa = 4.37AADD170 pKa = 4.9DD171 pKa = 4.37PSTEE175 pKa = 4.11ATDD178 pKa = 4.23DD179 pKa = 4.33PSDD182 pKa = 3.55SGYY185 pKa = 11.2SDD187 pKa = 3.55DD188 pKa = 4.02TGGGSEE194 pKa = 4.38EE195 pKa = 4.48APPEE199 pKa = 4.04PSVEE203 pKa = 4.29GQWQDD208 pKa = 3.55ADD210 pKa = 3.86AKK212 pKa = 9.52TLTVGTKK219 pKa = 8.33YY220 pKa = 9.53TSGDD224 pKa = 3.44LKK226 pKa = 11.03GKK228 pKa = 10.0YY229 pKa = 9.29ALSYY233 pKa = 10.72IDD235 pKa = 3.55TTGKK239 pKa = 10.87GILTGLGAYY248 pKa = 10.0RR249 pKa = 11.84DD250 pKa = 4.01DD251 pKa = 4.41DD252 pKa = 3.92SFRR255 pKa = 11.84LVLRR259 pKa = 11.84PMDD262 pKa = 4.45SDD264 pKa = 4.11SKK266 pKa = 11.55DD267 pKa = 3.4EE268 pKa = 3.99SDD270 pKa = 3.73YY271 pKa = 11.57VYY273 pKa = 10.29GTVRR277 pKa = 11.84RR278 pKa = 11.84SGDD281 pKa = 3.43DD282 pKa = 4.18VVITWDD288 pKa = 3.75DD289 pKa = 3.55GGKK292 pKa = 8.15EE293 pKa = 3.87TLGYY297 pKa = 10.05IGSLSDD303 pKa = 3.22
Molecular weight: 30.73 kDa
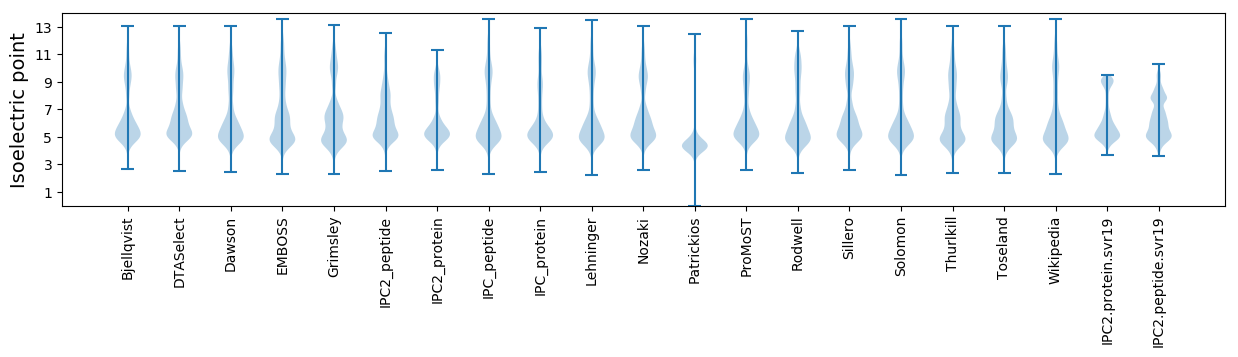
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5N8VWB1|A0A5N8VWB1_9ACTN Transposase family protein OS=Streptomyces phyllanthi OX=1803180 GN=FNH04_03235 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILASRR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.75GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILASRR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.75GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
Molecular weight: 5.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2865135 |
20 |
3672 |
331.7 |
35.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.011 ± 0.04 | 0.799 ± 0.007 |
6.063 ± 0.02 | 5.855 ± 0.025 |
2.759 ± 0.015 | 9.462 ± 0.028 |
2.313 ± 0.012 | 3.165 ± 0.017 |
2.191 ± 0.024 | 10.202 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.72 ± 0.01 | 1.861 ± 0.016 |
6.028 ± 0.02 | 2.746 ± 0.016 |
8.088 ± 0.032 | 5.258 ± 0.02 |
6.279 ± 0.025 | 8.484 ± 0.026 |
1.552 ± 0.011 | 2.164 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
