
Human gemycircularvirus GeTz1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemykibivirus; Gemykibivirus humas4; Human associated gemykibivirus 4
Average proteome isoelectric point is 6.47
Get precalculated fractions of proteins
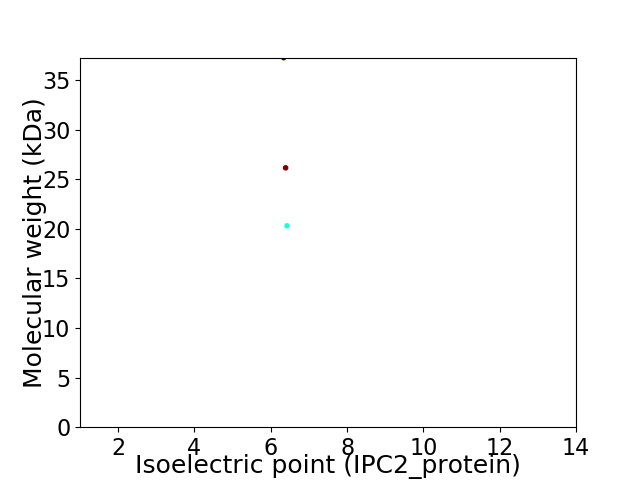
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A125R3J9|A0A125R3J9_9VIRU Uncharacterized protein OS=Human gemycircularvirus GeTz1 OX=1792832 PE=4 SV=1
MM1 pKa = 7.67SSFKK5 pKa = 10.61FQSRR9 pKa = 11.84YY10 pKa = 10.0VLLTYY15 pKa = 7.32PQCGDD20 pKa = 3.73LDD22 pKa = 3.62PWAVSDD28 pKa = 4.22HH29 pKa = 6.39LSSPRR34 pKa = 11.84AEE36 pKa = 4.25CIVGRR41 pKa = 11.84EE42 pKa = 3.92VHH44 pKa = 6.69RR45 pKa = 11.84DD46 pKa = 3.27GGIHH50 pKa = 5.95LHH52 pKa = 6.54CFADD56 pKa = 4.13FGKK59 pKa = 10.45RR60 pKa = 11.84FSSRR64 pKa = 11.84NTKK67 pKa = 9.62IFDD70 pKa = 3.7VGGHH74 pKa = 6.45HH75 pKa = 7.52PNTSPSKK82 pKa = 8.91GRR84 pKa = 11.84PGVGYY89 pKa = 10.3DD90 pKa = 3.44YY91 pKa = 10.78AIKK94 pKa = 10.9DD95 pKa = 3.49GDD97 pKa = 4.01VVAEE101 pKa = 4.03GLGRR105 pKa = 11.84PGGDD109 pKa = 3.02GDD111 pKa = 4.71QEE113 pKa = 4.19KK114 pKa = 10.8LPTNAEE120 pKa = 3.46RR121 pKa = 11.84WAEE124 pKa = 3.96IVAAEE129 pKa = 4.07SAEE132 pKa = 4.07EE133 pKa = 3.63FLEE136 pKa = 4.55YY137 pKa = 9.94YY138 pKa = 9.04WRR140 pKa = 11.84RR141 pKa = 11.84LDD143 pKa = 3.35PSAMVRR149 pKa = 11.84SFTQCRR155 pKa = 11.84AIRR158 pKa = 11.84RR159 pKa = 11.84ASLPSTFRR167 pKa = 11.84KK168 pKa = 6.91PTKK171 pKa = 10.26FSDD174 pKa = 3.07RR175 pKa = 11.84SIIRR179 pKa = 11.84NLQEE183 pKa = 3.35WRR185 pKa = 11.84DD186 pKa = 3.8SVNGLKK192 pKa = 10.6VILWGHH198 pKa = 5.33TGGGKK203 pKa = 10.31LSCPALATSLVLWGPSRR220 pKa = 11.84LGGTLWARR228 pKa = 11.84SLGNHH233 pKa = 7.12AYY235 pKa = 10.38FGGLFSMEE243 pKa = 3.77EE244 pKa = 4.43DD245 pKa = 3.21ITNVGYY251 pKa = 10.81AVFDD255 pKa = 4.95DD256 pKa = 3.5IGGLKK261 pKa = 10.18FLSTYY266 pKa = 10.55KK267 pKa = 10.47FWLSHH272 pKa = 4.5QKK274 pKa = 10.24EE275 pKa = 4.64FYY277 pKa = 9.5VTDD280 pKa = 3.94RR281 pKa = 11.84YY282 pKa = 10.46KK283 pKa = 11.02GKK285 pKa = 10.69KK286 pKa = 8.27LVQWGEE292 pKa = 3.48PAIWVNNTDD301 pKa = 3.03PRR303 pKa = 11.84EE304 pKa = 3.86EE305 pKa = 3.82HH306 pKa = 6.87GIRR309 pKa = 11.84DD310 pKa = 4.61EE311 pKa = 4.55EE312 pKa = 4.72IEE314 pKa = 4.15WLNANCQFVYY324 pKa = 10.15IGEE327 pKa = 4.41SIII330 pKa = 4.25
MM1 pKa = 7.67SSFKK5 pKa = 10.61FQSRR9 pKa = 11.84YY10 pKa = 10.0VLLTYY15 pKa = 7.32PQCGDD20 pKa = 3.73LDD22 pKa = 3.62PWAVSDD28 pKa = 4.22HH29 pKa = 6.39LSSPRR34 pKa = 11.84AEE36 pKa = 4.25CIVGRR41 pKa = 11.84EE42 pKa = 3.92VHH44 pKa = 6.69RR45 pKa = 11.84DD46 pKa = 3.27GGIHH50 pKa = 5.95LHH52 pKa = 6.54CFADD56 pKa = 4.13FGKK59 pKa = 10.45RR60 pKa = 11.84FSSRR64 pKa = 11.84NTKK67 pKa = 9.62IFDD70 pKa = 3.7VGGHH74 pKa = 6.45HH75 pKa = 7.52PNTSPSKK82 pKa = 8.91GRR84 pKa = 11.84PGVGYY89 pKa = 10.3DD90 pKa = 3.44YY91 pKa = 10.78AIKK94 pKa = 10.9DD95 pKa = 3.49GDD97 pKa = 4.01VVAEE101 pKa = 4.03GLGRR105 pKa = 11.84PGGDD109 pKa = 3.02GDD111 pKa = 4.71QEE113 pKa = 4.19KK114 pKa = 10.8LPTNAEE120 pKa = 3.46RR121 pKa = 11.84WAEE124 pKa = 3.96IVAAEE129 pKa = 4.07SAEE132 pKa = 4.07EE133 pKa = 3.63FLEE136 pKa = 4.55YY137 pKa = 9.94YY138 pKa = 9.04WRR140 pKa = 11.84RR141 pKa = 11.84LDD143 pKa = 3.35PSAMVRR149 pKa = 11.84SFTQCRR155 pKa = 11.84AIRR158 pKa = 11.84RR159 pKa = 11.84ASLPSTFRR167 pKa = 11.84KK168 pKa = 6.91PTKK171 pKa = 10.26FSDD174 pKa = 3.07RR175 pKa = 11.84SIIRR179 pKa = 11.84NLQEE183 pKa = 3.35WRR185 pKa = 11.84DD186 pKa = 3.8SVNGLKK192 pKa = 10.6VILWGHH198 pKa = 5.33TGGGKK203 pKa = 10.31LSCPALATSLVLWGPSRR220 pKa = 11.84LGGTLWARR228 pKa = 11.84SLGNHH233 pKa = 7.12AYY235 pKa = 10.38FGGLFSMEE243 pKa = 3.77EE244 pKa = 4.43DD245 pKa = 3.21ITNVGYY251 pKa = 10.81AVFDD255 pKa = 4.95DD256 pKa = 3.5IGGLKK261 pKa = 10.18FLSTYY266 pKa = 10.55KK267 pKa = 10.47FWLSHH272 pKa = 4.5QKK274 pKa = 10.24EE275 pKa = 4.64FYY277 pKa = 9.5VTDD280 pKa = 3.94RR281 pKa = 11.84YY282 pKa = 10.46KK283 pKa = 11.02GKK285 pKa = 10.69KK286 pKa = 8.27LVQWGEE292 pKa = 3.48PAIWVNNTDD301 pKa = 3.03PRR303 pKa = 11.84EE304 pKa = 3.86EE305 pKa = 3.82HH306 pKa = 6.87GIRR309 pKa = 11.84DD310 pKa = 4.61EE311 pKa = 4.55EE312 pKa = 4.72IEE314 pKa = 4.15WLNANCQFVYY324 pKa = 10.15IGEE327 pKa = 4.41SIII330 pKa = 4.25
Molecular weight: 37.25 kDa
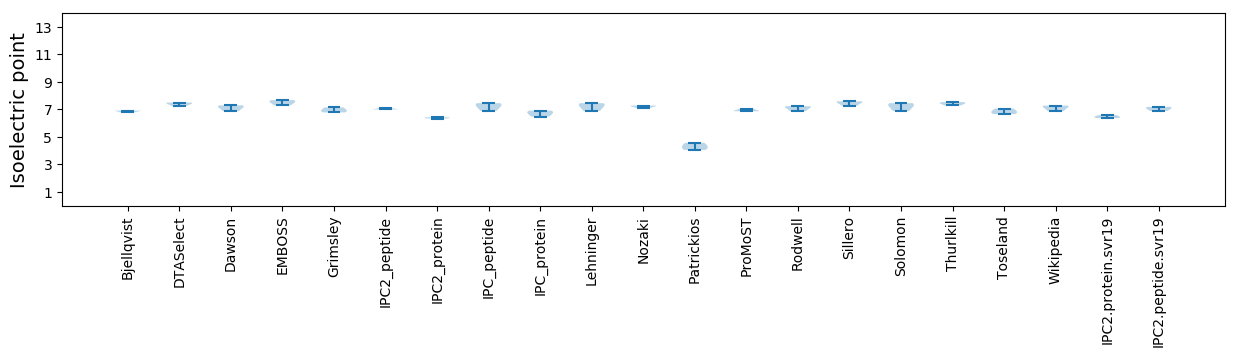
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A125R3J9|A0A125R3J9_9VIRU Uncharacterized protein OS=Human gemycircularvirus GeTz1 OX=1792832 PE=4 SV=1
MM1 pKa = 7.5SCLPTPNVEE10 pKa = 4.24TSTHH14 pKa = 5.67GLSLTTYY21 pKa = 9.84RR22 pKa = 11.84RR23 pKa = 11.84HH24 pKa = 5.8EE25 pKa = 4.32LSASWEE31 pKa = 4.19EE32 pKa = 4.21KK33 pKa = 9.96FIAMAEE39 pKa = 4.17FTSTVLQTLEE49 pKa = 3.98NDD51 pKa = 3.96SVRR54 pKa = 11.84GTQKK58 pKa = 10.33YY59 pKa = 10.57SMWEE63 pKa = 3.78DD64 pKa = 4.21TIRR67 pKa = 11.84TPALARR73 pKa = 11.84AGLGLDD79 pKa = 4.26MIMQSKK85 pKa = 9.14MEE87 pKa = 4.14MLSLKK92 pKa = 10.72DD93 pKa = 3.38LVAPEE98 pKa = 3.97EE99 pKa = 4.15MGTRR103 pKa = 11.84RR104 pKa = 11.84SFQQMLRR111 pKa = 11.84DD112 pKa = 4.04GLKK115 pKa = 10.23LWLQKK120 pKa = 8.53VQKK123 pKa = 10.23SFWSTIGGDD132 pKa = 3.58WILVRR137 pKa = 11.84WYY139 pKa = 10.21DD140 pKa = 3.48PSHH143 pKa = 6.33NAEE146 pKa = 4.02QYY148 pKa = 10.08AEE150 pKa = 3.88HH151 pKa = 7.7RR152 pKa = 11.84YY153 pKa = 9.85RR154 pKa = 11.84PLSGNLQNSPTGVLFGTFKK173 pKa = 11.02SGGTQQ178 pKa = 2.85
MM1 pKa = 7.5SCLPTPNVEE10 pKa = 4.24TSTHH14 pKa = 5.67GLSLTTYY21 pKa = 9.84RR22 pKa = 11.84RR23 pKa = 11.84HH24 pKa = 5.8EE25 pKa = 4.32LSASWEE31 pKa = 4.19EE32 pKa = 4.21KK33 pKa = 9.96FIAMAEE39 pKa = 4.17FTSTVLQTLEE49 pKa = 3.98NDD51 pKa = 3.96SVRR54 pKa = 11.84GTQKK58 pKa = 10.33YY59 pKa = 10.57SMWEE63 pKa = 3.78DD64 pKa = 4.21TIRR67 pKa = 11.84TPALARR73 pKa = 11.84AGLGLDD79 pKa = 4.26MIMQSKK85 pKa = 9.14MEE87 pKa = 4.14MLSLKK92 pKa = 10.72DD93 pKa = 3.38LVAPEE98 pKa = 3.97EE99 pKa = 4.15MGTRR103 pKa = 11.84RR104 pKa = 11.84SFQQMLRR111 pKa = 11.84DD112 pKa = 4.04GLKK115 pKa = 10.23LWLQKK120 pKa = 8.53VQKK123 pKa = 10.23SFWSTIGGDD132 pKa = 3.58WILVRR137 pKa = 11.84WYY139 pKa = 10.21DD140 pKa = 3.48PSHH143 pKa = 6.33NAEE146 pKa = 4.02QYY148 pKa = 10.08AEE150 pKa = 3.88HH151 pKa = 7.7RR152 pKa = 11.84YY153 pKa = 9.85RR154 pKa = 11.84PLSGNLQNSPTGVLFGTFKK173 pKa = 11.02SGGTQQ178 pKa = 2.85
Molecular weight: 20.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
744 |
178 |
330 |
248.0 |
27.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.317 ± 0.546 | 1.21 ± 0.326 |
5.914 ± 0.656 | 5.242 ± 1.077 |
4.032 ± 0.611 | 8.737 ± 0.835 |
2.419 ± 0.354 | 4.704 ± 0.575 |
4.704 ± 0.081 | 7.527 ± 1.437 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.285 ± 0.905 | 4.839 ± 1.45 |
3.763 ± 0.522 | 3.36 ± 0.701 |
6.317 ± 0.573 | 8.871 ± 0.535 |
7.527 ± 1.837 | 5.242 ± 0.403 |
3.091 ± 0.219 | 3.898 ± 0.49 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
