
Luteimonas marina
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Xanthomonadales; Xanthomonadaceae; Luteimonas
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
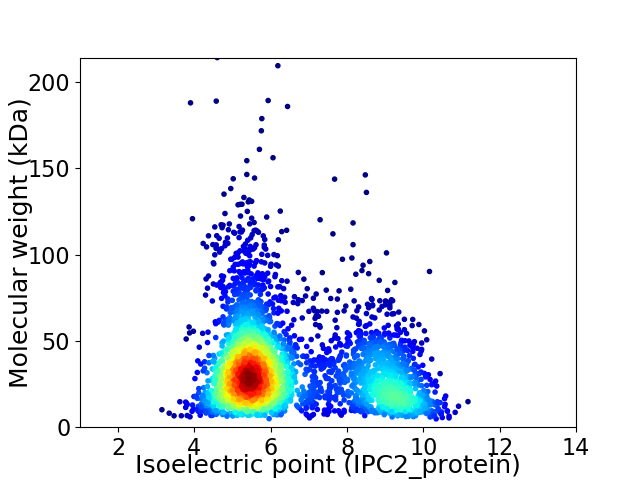
Virtual 2D-PAGE plot for 3401 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5C5U2P4|A0A5C5U2P4_9GAMM Zinc metalloprotease OS=Luteimonas marina OX=488485 GN=rseP PE=3 SV=1
MM1 pKa = 7.07NRR3 pKa = 11.84IYY5 pKa = 10.86CKK7 pKa = 10.27VWNQSKK13 pKa = 10.07GQVTVASEE21 pKa = 3.85LAKK24 pKa = 10.87SKK26 pKa = 11.03GKK28 pKa = 10.91GKK30 pKa = 10.06GARR33 pKa = 11.84LLQAVVVGGAIAFAGAAAAHH53 pKa = 6.35GPGHH57 pKa = 5.49ATKK60 pKa = 9.95PGSKK64 pKa = 9.78GQQASKK70 pKa = 10.52SQAHH74 pKa = 6.19GGKK77 pKa = 8.46PHH79 pKa = 5.19VHH81 pKa = 6.49EE82 pKa = 4.82KK83 pKa = 10.96LPAKK87 pKa = 10.46VVDD90 pKa = 3.78ACGTAVASATGAMACGNDD108 pKa = 3.47AAASGAGSTAVGNAAKK124 pKa = 8.97ATHH127 pKa = 6.64ANATALGNNARR138 pKa = 11.84AQQSNATAVGSNALASGSAATALGGATQAIGLRR171 pKa = 11.84STAVGYY177 pKa = 9.62SAQAHH182 pKa = 6.07AEE184 pKa = 4.0GAIALGFNAYY194 pKa = 9.44VGPAATGSVALGYY207 pKa = 9.68GARR210 pKa = 11.84TTEE213 pKa = 3.96ANVFSVGNGGGAGTVGPATRR233 pKa = 11.84RR234 pKa = 11.84ITNVSDD240 pKa = 5.03GIADD244 pKa = 3.43SDD246 pKa = 4.03AATVGQVGEE255 pKa = 4.21VADD258 pKa = 4.21AVDD261 pKa = 4.39AVAEE265 pKa = 4.25LAEE268 pKa = 4.24HH269 pKa = 6.22TDD271 pKa = 3.38RR272 pKa = 11.84YY273 pKa = 10.25FKK275 pKa = 11.18ADD277 pKa = 3.48GADD280 pKa = 3.98DD281 pKa = 4.8GSDD284 pKa = 3.45DD285 pKa = 4.9ASISGDD291 pKa = 3.42GAVAAGAGATADD303 pKa = 3.86GAGATALGAGSQATNAYY320 pKa = 7.17ATAVGNEE327 pKa = 3.81ASASGLQSTAAGFRR341 pKa = 11.84SDD343 pKa = 3.47ASGDD347 pKa = 3.32ASSAYY352 pKa = 9.98GGYY355 pKa = 9.97SVASGFGTSAFGYY368 pKa = 9.66GAEE371 pKa = 4.27AVLDD375 pKa = 4.03GATALGFGAVAGGYY389 pKa = 10.48DD390 pKa = 3.45SVAVGEE396 pKa = 4.41SALASGEE403 pKa = 3.87NSVAVGGAFWGLLPTEE419 pKa = 4.27AAGDD423 pKa = 3.76YY424 pKa = 8.55STAVGGAAYY433 pKa = 10.13AGGFNSTAIGNFSSAEE449 pKa = 4.0GDD451 pKa = 3.28SSLALGSDD459 pKa = 3.56SVALGEE465 pKa = 4.31GSVALGQGAYY475 pKa = 10.29ADD477 pKa = 3.73RR478 pKa = 11.84DD479 pKa = 3.97YY480 pKa = 10.85TVSVGDD486 pKa = 3.55VGAEE490 pKa = 3.64RR491 pKa = 11.84QITNVAAGTEE501 pKa = 4.26GTDD504 pKa = 3.83AVNVDD509 pKa = 3.65QLEE512 pKa = 4.08EE513 pKa = 3.96ATASNRR519 pKa = 11.84YY520 pKa = 8.0FQATGNDD527 pKa = 4.31DD528 pKa = 4.67DD529 pKa = 5.52PSDD532 pKa = 4.27DD533 pKa = 3.34VGAYY537 pKa = 10.31AEE539 pKa = 4.34GAYY542 pKa = 8.82ATASGEE548 pKa = 3.97ATNAIGQGASAYY560 pKa = 10.42GSGAYY565 pKa = 9.97AEE567 pKa = 4.75GLNATASGYY576 pKa = 10.47NATATADD583 pKa = 3.26GATAVGGSLYY593 pKa = 11.03YY594 pKa = 10.27EE595 pKa = 4.91DD596 pKa = 4.35PTTGEE601 pKa = 4.11VLLDD605 pKa = 3.47QTTTASAVGASAYY618 pKa = 10.36GAGAQANGAFSTASGAAATADD639 pKa = 3.88GLQSSASGYY648 pKa = 10.62SSAASGDD655 pKa = 3.69YY656 pKa = 7.7TTATGSFSNAAGYY669 pKa = 10.08GGSALGYY676 pKa = 9.18GAEE679 pKa = 4.18ASGDD683 pKa = 3.57YY684 pKa = 9.18ATAVGVVATASGVSSVAVGEE704 pKa = 4.24FSEE707 pKa = 4.45ATGDD711 pKa = 3.44EE712 pKa = 4.47SVAVGGSTFFGFIPAQASGTGATAVGAGAWATGDD746 pKa = 3.52YY747 pKa = 8.55GTALGWNSWANAEE760 pKa = 4.27SATALGEE767 pKa = 4.27SATASAANSVALGAGSLADD786 pKa = 3.92RR787 pKa = 11.84EE788 pKa = 4.54NTVSVGSAGAEE799 pKa = 3.46RR800 pKa = 11.84QVTNVAAGTEE810 pKa = 4.1ATDD813 pKa = 4.26AVNLEE818 pKa = 4.09QLEE821 pKa = 4.39EE822 pKa = 4.26ATQHH826 pKa = 4.77TRR828 pKa = 11.84YY829 pKa = 9.15FAASGGADD837 pKa = 3.77SDD839 pKa = 3.44NGAYY843 pKa = 10.25VEE845 pKa = 4.42GDD847 pKa = 3.52YY848 pKa = 10.57ATAAGEE854 pKa = 4.12SATAVGEE861 pKa = 4.31GASALGSGAFALADD875 pKa = 3.44YY876 pKa = 8.05ATAVGFNAVADD887 pKa = 3.9QASAVAVGASSTALGEE903 pKa = 4.02YY904 pKa = 9.97AVAVGSEE911 pKa = 4.1SFAEE915 pKa = 4.98GFASTATGAAAMAVGEE931 pKa = 4.32GSTAAGALATASGIEE946 pKa = 4.2STAVGAFAEE955 pKa = 4.45ATGDD959 pKa = 3.56LATAAGAEE967 pKa = 4.51SVASGNEE974 pKa = 3.73SSAFGALSEE983 pKa = 4.47ASGHH987 pKa = 5.63YY988 pKa = 8.36ATAVGARR995 pKa = 11.84ADD997 pKa = 3.33ASGFNSTSLGSWSTASGFNATAVGADD1023 pKa = 3.62SVASGDD1029 pKa = 3.71DD1030 pKa = 3.51SLAVGQAAIASGAEE1044 pKa = 4.12SVAVGGSSLSGFFPTEE1060 pKa = 3.34ASGDD1064 pKa = 3.67YY1065 pKa = 9.2ATAVGAGAWAVGTNSTALGNFATAEE1090 pKa = 4.44GEE1092 pKa = 4.04DD1093 pKa = 3.87STALGVGSYY1102 pKa = 10.91ASADD1106 pKa = 3.35NSVALGVGSEE1116 pKa = 4.15ADD1118 pKa = 3.57RR1119 pKa = 11.84EE1120 pKa = 4.74GTVSVGSVGSEE1131 pKa = 3.5RR1132 pKa = 11.84QITNVAAGTEE1142 pKa = 4.23GTDD1145 pKa = 4.01AVNLEE1150 pKa = 4.07QLEE1153 pKa = 4.23EE1154 pKa = 4.2ATASTRR1160 pKa = 11.84YY1161 pKa = 8.8FQATGNDD1168 pKa = 4.31DD1169 pKa = 4.67DD1170 pKa = 5.52PSDD1173 pKa = 4.27DD1174 pKa = 3.34VGAYY1178 pKa = 10.26AEE1180 pKa = 4.4GTYY1183 pKa = 9.32ATASGEE1189 pKa = 3.99ATNAIGQGASAYY1201 pKa = 10.42GSGAYY1206 pKa = 9.97AEE1208 pKa = 4.75GLNATASGYY1217 pKa = 10.47NATATADD1224 pKa = 3.26GATAVGGSLYY1234 pKa = 11.03YY1235 pKa = 10.27EE1236 pKa = 4.91DD1237 pKa = 4.35PTTGEE1242 pKa = 4.11VLLDD1246 pKa = 3.47QTTTASAVGASAFGAGAQANGAFSTASGAAATADD1280 pKa = 3.75GLQATAIGYY1289 pKa = 9.94SSTSSGLAATSLGGFSEE1306 pKa = 4.31ATGDD1310 pKa = 3.46FSTALGYY1317 pKa = 10.21GAAASNTRR1325 pKa = 11.84TTAVGITSTASGLNATAVGNTSTASGNASAALGSSSNATGLNATALGASAWATADD1380 pKa = 3.2NATSVGQLAWATVAGGTAIGRR1401 pKa = 11.84DD1402 pKa = 3.49AYY1404 pKa = 11.38VYY1406 pKa = 10.5GAGVNATAVGLSAWANGASSVALGAGARR1434 pKa = 11.84ATEE1437 pKa = 4.18EE1438 pKa = 4.08NVVSVGNGTGAGGHH1452 pKa = 6.41PATRR1456 pKa = 11.84RR1457 pKa = 11.84IVNVGDD1463 pKa = 3.64GVAASDD1469 pKa = 3.66AATVGQLEE1477 pKa = 4.24AVTEE1481 pKa = 4.11HH1482 pKa = 5.51TRR1484 pKa = 11.84YY1485 pKa = 9.17FAASGGADD1493 pKa = 3.67SDD1495 pKa = 3.59NAAYY1499 pKa = 10.84VEE1501 pKa = 4.17GDD1503 pKa = 3.57YY1504 pKa = 10.55ATAAGEE1510 pKa = 4.12SATAVGEE1517 pKa = 4.31GASALGSGAFALAEE1531 pKa = 3.92RR1532 pKa = 11.84ATAVGFNASAGGVDD1546 pKa = 4.3AVALGASASAIGDD1559 pKa = 3.56GSVVLGAGAVSWGSGAVAVGSGGTANADD1587 pKa = 2.98AATAVGGFANAAGTNSVAVGSGAVSLGDD1615 pKa = 3.44HH1616 pKa = 5.72STALGADD1623 pKa = 3.73AFAEE1627 pKa = 4.27ADD1629 pKa = 3.62GSVALGQGSVADD1641 pKa = 4.17RR1642 pKa = 11.84ANTVSVGSAGSEE1654 pKa = 3.63RR1655 pKa = 11.84QITNVAAGRR1664 pKa = 11.84IALGSTDD1671 pKa = 4.07AVTGGQVYY1679 pKa = 10.33DD1680 pKa = 4.54ALDD1683 pKa = 3.43STAQILGGGAGVTAFGTLSAPAYY1706 pKa = 7.68MIQGRR1711 pKa = 11.84TYY1713 pKa = 10.6FSVGDD1718 pKa = 3.52AFGAVDD1724 pKa = 3.69AQISVLGNRR1733 pKa = 11.84LNSLEE1738 pKa = 4.15ASSMASEE1745 pKa = 4.48NSTASAAASRR1755 pKa = 11.84RR1756 pKa = 11.84TGNGIDD1762 pKa = 4.43NGDD1765 pKa = 3.58ATDD1768 pKa = 3.86GGLVHH1773 pKa = 7.46DD1774 pKa = 5.58RR1775 pKa = 11.84GTDD1778 pKa = 3.31TVGSGRR1784 pKa = 11.84AGGTQVVDD1792 pKa = 3.7TATSTAVTVGQGAVATQATATAIGDD1817 pKa = 3.94GATATAAGSVALGQGSVADD1836 pKa = 4.17RR1837 pKa = 11.84ANTVSVGSAGNEE1849 pKa = 3.59RR1850 pKa = 11.84QITNVAAGTQSTDD1863 pKa = 3.02AVNVAQLEE1871 pKa = 4.56STATQAVADD1880 pKa = 4.25ANAYY1884 pKa = 8.11TDD1886 pKa = 3.48QKK1888 pKa = 10.78FSAWSDD1894 pKa = 3.38SFSAYY1899 pKa = 7.38QTQLEE1904 pKa = 4.28QRR1906 pKa = 11.84FTDD1909 pKa = 3.04QDD1911 pKa = 3.22RR1912 pKa = 11.84RR1913 pKa = 11.84IDD1915 pKa = 3.45RR1916 pKa = 11.84QGAMGAAMLNMATSAAGIRR1935 pKa = 11.84TQNRR1939 pKa = 11.84VGVGVGFQGGQSALSLGYY1957 pKa = 10.06QRR1959 pKa = 11.84AISDD1963 pKa = 3.79RR1964 pKa = 11.84ATLTLGGAFSGDD1976 pKa = 3.49EE1977 pKa = 3.95KK1978 pKa = 11.26SVGVGAGFGWW1988 pKa = 3.55
MM1 pKa = 7.07NRR3 pKa = 11.84IYY5 pKa = 10.86CKK7 pKa = 10.27VWNQSKK13 pKa = 10.07GQVTVASEE21 pKa = 3.85LAKK24 pKa = 10.87SKK26 pKa = 11.03GKK28 pKa = 10.91GKK30 pKa = 10.06GARR33 pKa = 11.84LLQAVVVGGAIAFAGAAAAHH53 pKa = 6.35GPGHH57 pKa = 5.49ATKK60 pKa = 9.95PGSKK64 pKa = 9.78GQQASKK70 pKa = 10.52SQAHH74 pKa = 6.19GGKK77 pKa = 8.46PHH79 pKa = 5.19VHH81 pKa = 6.49EE82 pKa = 4.82KK83 pKa = 10.96LPAKK87 pKa = 10.46VVDD90 pKa = 3.78ACGTAVASATGAMACGNDD108 pKa = 3.47AAASGAGSTAVGNAAKK124 pKa = 8.97ATHH127 pKa = 6.64ANATALGNNARR138 pKa = 11.84AQQSNATAVGSNALASGSAATALGGATQAIGLRR171 pKa = 11.84STAVGYY177 pKa = 9.62SAQAHH182 pKa = 6.07AEE184 pKa = 4.0GAIALGFNAYY194 pKa = 9.44VGPAATGSVALGYY207 pKa = 9.68GARR210 pKa = 11.84TTEE213 pKa = 3.96ANVFSVGNGGGAGTVGPATRR233 pKa = 11.84RR234 pKa = 11.84ITNVSDD240 pKa = 5.03GIADD244 pKa = 3.43SDD246 pKa = 4.03AATVGQVGEE255 pKa = 4.21VADD258 pKa = 4.21AVDD261 pKa = 4.39AVAEE265 pKa = 4.25LAEE268 pKa = 4.24HH269 pKa = 6.22TDD271 pKa = 3.38RR272 pKa = 11.84YY273 pKa = 10.25FKK275 pKa = 11.18ADD277 pKa = 3.48GADD280 pKa = 3.98DD281 pKa = 4.8GSDD284 pKa = 3.45DD285 pKa = 4.9ASISGDD291 pKa = 3.42GAVAAGAGATADD303 pKa = 3.86GAGATALGAGSQATNAYY320 pKa = 7.17ATAVGNEE327 pKa = 3.81ASASGLQSTAAGFRR341 pKa = 11.84SDD343 pKa = 3.47ASGDD347 pKa = 3.32ASSAYY352 pKa = 9.98GGYY355 pKa = 9.97SVASGFGTSAFGYY368 pKa = 9.66GAEE371 pKa = 4.27AVLDD375 pKa = 4.03GATALGFGAVAGGYY389 pKa = 10.48DD390 pKa = 3.45SVAVGEE396 pKa = 4.41SALASGEE403 pKa = 3.87NSVAVGGAFWGLLPTEE419 pKa = 4.27AAGDD423 pKa = 3.76YY424 pKa = 8.55STAVGGAAYY433 pKa = 10.13AGGFNSTAIGNFSSAEE449 pKa = 4.0GDD451 pKa = 3.28SSLALGSDD459 pKa = 3.56SVALGEE465 pKa = 4.31GSVALGQGAYY475 pKa = 10.29ADD477 pKa = 3.73RR478 pKa = 11.84DD479 pKa = 3.97YY480 pKa = 10.85TVSVGDD486 pKa = 3.55VGAEE490 pKa = 3.64RR491 pKa = 11.84QITNVAAGTEE501 pKa = 4.26GTDD504 pKa = 3.83AVNVDD509 pKa = 3.65QLEE512 pKa = 4.08EE513 pKa = 3.96ATASNRR519 pKa = 11.84YY520 pKa = 8.0FQATGNDD527 pKa = 4.31DD528 pKa = 4.67DD529 pKa = 5.52PSDD532 pKa = 4.27DD533 pKa = 3.34VGAYY537 pKa = 10.31AEE539 pKa = 4.34GAYY542 pKa = 8.82ATASGEE548 pKa = 3.97ATNAIGQGASAYY560 pKa = 10.42GSGAYY565 pKa = 9.97AEE567 pKa = 4.75GLNATASGYY576 pKa = 10.47NATATADD583 pKa = 3.26GATAVGGSLYY593 pKa = 11.03YY594 pKa = 10.27EE595 pKa = 4.91DD596 pKa = 4.35PTTGEE601 pKa = 4.11VLLDD605 pKa = 3.47QTTTASAVGASAYY618 pKa = 10.36GAGAQANGAFSTASGAAATADD639 pKa = 3.88GLQSSASGYY648 pKa = 10.62SSAASGDD655 pKa = 3.69YY656 pKa = 7.7TTATGSFSNAAGYY669 pKa = 10.08GGSALGYY676 pKa = 9.18GAEE679 pKa = 4.18ASGDD683 pKa = 3.57YY684 pKa = 9.18ATAVGVVATASGVSSVAVGEE704 pKa = 4.24FSEE707 pKa = 4.45ATGDD711 pKa = 3.44EE712 pKa = 4.47SVAVGGSTFFGFIPAQASGTGATAVGAGAWATGDD746 pKa = 3.52YY747 pKa = 8.55GTALGWNSWANAEE760 pKa = 4.27SATALGEE767 pKa = 4.27SATASAANSVALGAGSLADD786 pKa = 3.92RR787 pKa = 11.84EE788 pKa = 4.54NTVSVGSAGAEE799 pKa = 3.46RR800 pKa = 11.84QVTNVAAGTEE810 pKa = 4.1ATDD813 pKa = 4.26AVNLEE818 pKa = 4.09QLEE821 pKa = 4.39EE822 pKa = 4.26ATQHH826 pKa = 4.77TRR828 pKa = 11.84YY829 pKa = 9.15FAASGGADD837 pKa = 3.77SDD839 pKa = 3.44NGAYY843 pKa = 10.25VEE845 pKa = 4.42GDD847 pKa = 3.52YY848 pKa = 10.57ATAAGEE854 pKa = 4.12SATAVGEE861 pKa = 4.31GASALGSGAFALADD875 pKa = 3.44YY876 pKa = 8.05ATAVGFNAVADD887 pKa = 3.9QASAVAVGASSTALGEE903 pKa = 4.02YY904 pKa = 9.97AVAVGSEE911 pKa = 4.1SFAEE915 pKa = 4.98GFASTATGAAAMAVGEE931 pKa = 4.32GSTAAGALATASGIEE946 pKa = 4.2STAVGAFAEE955 pKa = 4.45ATGDD959 pKa = 3.56LATAAGAEE967 pKa = 4.51SVASGNEE974 pKa = 3.73SSAFGALSEE983 pKa = 4.47ASGHH987 pKa = 5.63YY988 pKa = 8.36ATAVGARR995 pKa = 11.84ADD997 pKa = 3.33ASGFNSTSLGSWSTASGFNATAVGADD1023 pKa = 3.62SVASGDD1029 pKa = 3.71DD1030 pKa = 3.51SLAVGQAAIASGAEE1044 pKa = 4.12SVAVGGSSLSGFFPTEE1060 pKa = 3.34ASGDD1064 pKa = 3.67YY1065 pKa = 9.2ATAVGAGAWAVGTNSTALGNFATAEE1090 pKa = 4.44GEE1092 pKa = 4.04DD1093 pKa = 3.87STALGVGSYY1102 pKa = 10.91ASADD1106 pKa = 3.35NSVALGVGSEE1116 pKa = 4.15ADD1118 pKa = 3.57RR1119 pKa = 11.84EE1120 pKa = 4.74GTVSVGSVGSEE1131 pKa = 3.5RR1132 pKa = 11.84QITNVAAGTEE1142 pKa = 4.23GTDD1145 pKa = 4.01AVNLEE1150 pKa = 4.07QLEE1153 pKa = 4.23EE1154 pKa = 4.2ATASTRR1160 pKa = 11.84YY1161 pKa = 8.8FQATGNDD1168 pKa = 4.31DD1169 pKa = 4.67DD1170 pKa = 5.52PSDD1173 pKa = 4.27DD1174 pKa = 3.34VGAYY1178 pKa = 10.26AEE1180 pKa = 4.4GTYY1183 pKa = 9.32ATASGEE1189 pKa = 3.99ATNAIGQGASAYY1201 pKa = 10.42GSGAYY1206 pKa = 9.97AEE1208 pKa = 4.75GLNATASGYY1217 pKa = 10.47NATATADD1224 pKa = 3.26GATAVGGSLYY1234 pKa = 11.03YY1235 pKa = 10.27EE1236 pKa = 4.91DD1237 pKa = 4.35PTTGEE1242 pKa = 4.11VLLDD1246 pKa = 3.47QTTTASAVGASAFGAGAQANGAFSTASGAAATADD1280 pKa = 3.75GLQATAIGYY1289 pKa = 9.94SSTSSGLAATSLGGFSEE1306 pKa = 4.31ATGDD1310 pKa = 3.46FSTALGYY1317 pKa = 10.21GAAASNTRR1325 pKa = 11.84TTAVGITSTASGLNATAVGNTSTASGNASAALGSSSNATGLNATALGASAWATADD1380 pKa = 3.2NATSVGQLAWATVAGGTAIGRR1401 pKa = 11.84DD1402 pKa = 3.49AYY1404 pKa = 11.38VYY1406 pKa = 10.5GAGVNATAVGLSAWANGASSVALGAGARR1434 pKa = 11.84ATEE1437 pKa = 4.18EE1438 pKa = 4.08NVVSVGNGTGAGGHH1452 pKa = 6.41PATRR1456 pKa = 11.84RR1457 pKa = 11.84IVNVGDD1463 pKa = 3.64GVAASDD1469 pKa = 3.66AATVGQLEE1477 pKa = 4.24AVTEE1481 pKa = 4.11HH1482 pKa = 5.51TRR1484 pKa = 11.84YY1485 pKa = 9.17FAASGGADD1493 pKa = 3.67SDD1495 pKa = 3.59NAAYY1499 pKa = 10.84VEE1501 pKa = 4.17GDD1503 pKa = 3.57YY1504 pKa = 10.55ATAAGEE1510 pKa = 4.12SATAVGEE1517 pKa = 4.31GASALGSGAFALAEE1531 pKa = 3.92RR1532 pKa = 11.84ATAVGFNASAGGVDD1546 pKa = 4.3AVALGASASAIGDD1559 pKa = 3.56GSVVLGAGAVSWGSGAVAVGSGGTANADD1587 pKa = 2.98AATAVGGFANAAGTNSVAVGSGAVSLGDD1615 pKa = 3.44HH1616 pKa = 5.72STALGADD1623 pKa = 3.73AFAEE1627 pKa = 4.27ADD1629 pKa = 3.62GSVALGQGSVADD1641 pKa = 4.17RR1642 pKa = 11.84ANTVSVGSAGSEE1654 pKa = 3.63RR1655 pKa = 11.84QITNVAAGRR1664 pKa = 11.84IALGSTDD1671 pKa = 4.07AVTGGQVYY1679 pKa = 10.33DD1680 pKa = 4.54ALDD1683 pKa = 3.43STAQILGGGAGVTAFGTLSAPAYY1706 pKa = 7.68MIQGRR1711 pKa = 11.84TYY1713 pKa = 10.6FSVGDD1718 pKa = 3.52AFGAVDD1724 pKa = 3.69AQISVLGNRR1733 pKa = 11.84LNSLEE1738 pKa = 4.15ASSMASEE1745 pKa = 4.48NSTASAAASRR1755 pKa = 11.84RR1756 pKa = 11.84TGNGIDD1762 pKa = 4.43NGDD1765 pKa = 3.58ATDD1768 pKa = 3.86GGLVHH1773 pKa = 7.46DD1774 pKa = 5.58RR1775 pKa = 11.84GTDD1778 pKa = 3.31TVGSGRR1784 pKa = 11.84AGGTQVVDD1792 pKa = 3.7TATSTAVTVGQGAVATQATATAIGDD1817 pKa = 3.94GATATAAGSVALGQGSVADD1836 pKa = 4.17RR1837 pKa = 11.84ANTVSVGSAGNEE1849 pKa = 3.59RR1850 pKa = 11.84QITNVAAGTQSTDD1863 pKa = 3.02AVNVAQLEE1871 pKa = 4.56STATQAVADD1880 pKa = 4.25ANAYY1884 pKa = 8.11TDD1886 pKa = 3.48QKK1888 pKa = 10.78FSAWSDD1894 pKa = 3.38SFSAYY1899 pKa = 7.38QTQLEE1904 pKa = 4.28QRR1906 pKa = 11.84FTDD1909 pKa = 3.04QDD1911 pKa = 3.22RR1912 pKa = 11.84RR1913 pKa = 11.84IDD1915 pKa = 3.45RR1916 pKa = 11.84QGAMGAAMLNMATSAAGIRR1935 pKa = 11.84TQNRR1939 pKa = 11.84VGVGVGFQGGQSALSLGYY1957 pKa = 10.06QRR1959 pKa = 11.84AISDD1963 pKa = 3.79RR1964 pKa = 11.84ATLTLGGAFSGDD1976 pKa = 3.49EE1977 pKa = 3.95KK1978 pKa = 11.26SVGVGAGFGWW1988 pKa = 3.55
Molecular weight: 188.06 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5C5UB04|A0A5C5UB04_9GAMM Protease 4 OS=Luteimonas marina OX=488485 GN=sppA PE=3 SV=1
MM1 pKa = 7.09TRR3 pKa = 11.84SATPRR8 pKa = 11.84PTAQADD14 pKa = 3.54ALRR17 pKa = 11.84TLEE20 pKa = 4.28HH21 pKa = 6.92LLLGALFLGAIAMLSLPAARR41 pKa = 11.84GASATLGWMPLWLLGLPAASLATAFALRR69 pKa = 11.84LSRR72 pKa = 11.84RR73 pKa = 11.84VAMQAPSSAPRR84 pKa = 11.84RR85 pKa = 11.84RR86 pKa = 11.84RR87 pKa = 11.84PATFAATRR95 pKa = 11.84RR96 pKa = 11.84RR97 pKa = 11.84TGAPRR102 pKa = 11.84RR103 pKa = 11.84ASRR106 pKa = 11.84LLAAIVLRR114 pKa = 4.48
MM1 pKa = 7.09TRR3 pKa = 11.84SATPRR8 pKa = 11.84PTAQADD14 pKa = 3.54ALRR17 pKa = 11.84TLEE20 pKa = 4.28HH21 pKa = 6.92LLLGALFLGAIAMLSLPAARR41 pKa = 11.84GASATLGWMPLWLLGLPAASLATAFALRR69 pKa = 11.84LSRR72 pKa = 11.84RR73 pKa = 11.84VAMQAPSSAPRR84 pKa = 11.84RR85 pKa = 11.84RR86 pKa = 11.84RR87 pKa = 11.84PATFAATRR95 pKa = 11.84RR96 pKa = 11.84RR97 pKa = 11.84TGAPRR102 pKa = 11.84RR103 pKa = 11.84ASRR106 pKa = 11.84LLAAIVLRR114 pKa = 4.48
Molecular weight: 12.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1148263 |
41 |
1988 |
337.6 |
36.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.489 ± 0.066 | 0.792 ± 0.012 |
6.188 ± 0.037 | 5.461 ± 0.036 |
3.395 ± 0.026 | 8.864 ± 0.037 |
2.216 ± 0.022 | 4.066 ± 0.03 |
2.496 ± 0.027 | 10.678 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.078 ± 0.019 | 2.294 ± 0.025 |
5.522 ± 0.032 | 3.383 ± 0.026 |
8.099 ± 0.048 | 4.961 ± 0.03 |
4.7 ± 0.029 | 7.5 ± 0.037 |
1.567 ± 0.017 | 2.25 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
