
Butyricicoccus pullicaecorum 1.2
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Butyricicoccus; Butyricicoccus pullicaecorum
Average proteome isoelectric point is 6.04
Get precalculated fractions of proteins
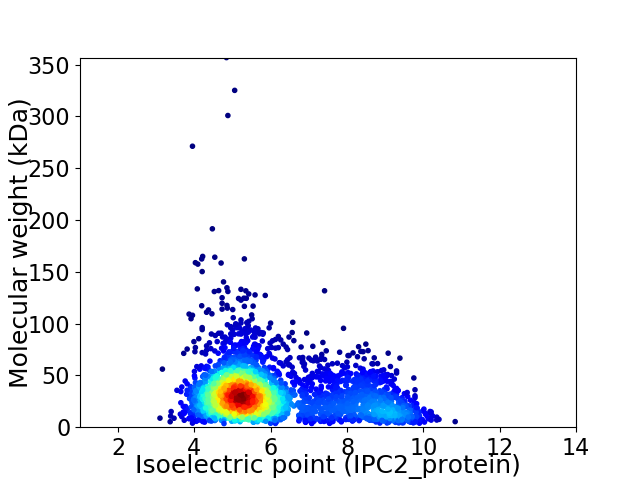
Virtual 2D-PAGE plot for 3107 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R8VUF9|R8VUF9_9CLOT Cys_rich_VLP domain-containing protein OS=Butyricicoccus pullicaecorum 1.2 OX=1203606 GN=HMPREF1526_02220 PE=4 SV=1
MM1 pKa = 7.8KK2 pKa = 10.25KK3 pKa = 10.24RR4 pKa = 11.84FWACFLSLSLVLTMMPTMAFAADD27 pKa = 4.03DD28 pKa = 4.06EE29 pKa = 4.97VNEE32 pKa = 4.27PGGTVEE38 pKa = 4.21VCVEE42 pKa = 4.21TEE44 pKa = 3.95GCTLEE49 pKa = 4.75AGHH52 pKa = 6.51EE53 pKa = 4.55GEE55 pKa = 4.53CVVVEE60 pKa = 4.4SEE62 pKa = 4.26PEE64 pKa = 4.0EE65 pKa = 4.64PEE67 pKa = 4.05SAPCKK72 pKa = 8.55VTEE75 pKa = 4.32GCTLEE80 pKa = 4.35NGHH83 pKa = 6.37EE84 pKa = 4.54GEE86 pKa = 4.51CVVAEE91 pKa = 4.41SEE93 pKa = 4.33PEE95 pKa = 3.94EE96 pKa = 4.62PEE98 pKa = 4.47SVPCTVTEE106 pKa = 4.35GCTLEE111 pKa = 5.22DD112 pKa = 3.59GHH114 pKa = 7.05EE115 pKa = 4.57GGCVVEE121 pKa = 4.9SSVDD125 pKa = 3.51SLSEE129 pKa = 4.01DD130 pKa = 3.61EE131 pKa = 5.23EE132 pKa = 4.36IVISITEE139 pKa = 3.96ADD141 pKa = 3.78MNSADD146 pKa = 4.8EE147 pKa = 4.39EE148 pKa = 4.31NPPIKK153 pKa = 10.39SAIARR158 pKa = 11.84LSLADD163 pKa = 4.02PSSVTQLKK171 pKa = 10.08VVTEE175 pKa = 4.31GEE177 pKa = 4.53AYY179 pKa = 10.75LSPEE183 pKa = 4.0DD184 pKa = 3.52NEE186 pKa = 4.47YY187 pKa = 10.67IKK189 pKa = 10.0TVFSSLNYY197 pKa = 9.97LDD199 pKa = 5.45EE200 pKa = 4.95SEE202 pKa = 4.89CQCSTRR208 pKa = 11.84KK209 pKa = 8.44LTEE212 pKa = 4.31AYY214 pKa = 9.75LGKK217 pKa = 10.46SGLPDD222 pKa = 3.05EE223 pKa = 5.66LYY225 pKa = 11.15AEE227 pKa = 4.63FGGLSGYY234 pKa = 9.26TSLEE238 pKa = 3.87TLILPEE244 pKa = 3.91EE245 pKa = 4.34TQVVNSYY252 pKa = 8.46TLSEE256 pKa = 4.27RR257 pKa = 11.84GGGVNDD263 pKa = 3.86TGLTEE268 pKa = 5.56LNLSNNILVIEE279 pKa = 4.39SGAFSGNTNFVGNLVIPDD297 pKa = 3.59SVVFIGNNSFGVGNEE312 pKa = 4.73LIACGTLTLGDD323 pKa = 3.79SVKK326 pKa = 11.13YY327 pKa = 10.23IDD329 pKa = 4.39ANGFKK334 pKa = 10.57SRR336 pKa = 11.84LFEE339 pKa = 4.89GDD341 pKa = 3.75LVIPNSVEE349 pKa = 4.13MINNWSFSPGAFKK362 pKa = 11.05NGTWTLGDD370 pKa = 3.48NLINIGSAALKK381 pKa = 9.43GICSGNSGTFIVLNQYY397 pKa = 10.66KK398 pKa = 9.63MSTQAFADD406 pKa = 3.49NSFEE410 pKa = 4.42KK411 pKa = 10.45IAFEE415 pKa = 4.73EE416 pKa = 5.46GITDD420 pKa = 3.54ISTSQVIASSKK431 pKa = 9.94NIEE434 pKa = 4.36EE435 pKa = 4.08IVLPSTLEE443 pKa = 4.32SISGDD448 pKa = 3.71SAFAGCPALTTINFPKK464 pKa = 10.41SLEE467 pKa = 4.07RR468 pKa = 11.84LGDD471 pKa = 3.45RR472 pKa = 11.84TFEE475 pKa = 4.26NDD477 pKa = 3.29DD478 pKa = 3.66ALTSVVLPSNLTQIGSFTFEE498 pKa = 4.2DD499 pKa = 3.8SGILGLYY506 pKa = 9.52IPEE509 pKa = 4.27SVTDD513 pKa = 3.21IGNRR517 pKa = 11.84IVQDD521 pKa = 3.48IPSGSIVYY529 pKa = 8.81VANEE533 pKa = 3.65ALLNMMNEE541 pKa = 4.74SDD543 pKa = 3.91SAWNNKK549 pKa = 8.03YY550 pKa = 10.84NNEE553 pKa = 3.92TTALAMTNGGSFKK566 pKa = 10.67EE567 pKa = 3.99DD568 pKa = 3.37TEE570 pKa = 4.43FSEE573 pKa = 5.47SVLATPIKK581 pKa = 10.2SGNIFAGWYY590 pKa = 9.96ADD592 pKa = 3.87EE593 pKa = 4.49EE594 pKa = 4.37LADD597 pKa = 3.9EE598 pKa = 4.6VVTNADD604 pKa = 3.4AGNTYY609 pKa = 9.4YY610 pKa = 11.13AEE612 pKa = 4.19WLPSNYY618 pKa = 9.25TVTEE622 pKa = 4.25NINFGSVVYY631 pKa = 10.39GSTASRR637 pKa = 11.84VIQVNGSDD645 pKa = 3.29VAGALKK651 pKa = 10.77VAGNDD656 pKa = 3.09IFDD659 pKa = 3.95TVVNEE664 pKa = 4.43DD665 pKa = 3.1NTITVTPHH673 pKa = 6.06SNLSVGEE680 pKa = 4.13YY681 pKa = 10.88EE682 pKa = 3.82EE683 pKa = 4.27TLYY686 pKa = 9.85VTTPDD691 pKa = 3.09NATFFVPVTLEE702 pKa = 3.95VVPASSKK709 pKa = 11.17LEE711 pKa = 3.52IWANGAKK718 pKa = 10.07KK719 pKa = 10.44LSLSGGGNVTLTITGDD735 pKa = 3.6LNADD739 pKa = 3.76EE740 pKa = 4.67VTVTCNNEE748 pKa = 4.1NIEE751 pKa = 4.25VTKK754 pKa = 11.0NADD757 pKa = 3.17DD758 pKa = 4.36TYY760 pKa = 11.21TVTLPNEE767 pKa = 3.98TADD770 pKa = 3.64YY771 pKa = 9.53TFTATYY777 pKa = 10.67DD778 pKa = 3.31GDD780 pKa = 4.15QNHH783 pKa = 5.92EE784 pKa = 4.22MARR787 pKa = 11.84ATCTVSVTRR796 pKa = 11.84HH797 pKa = 4.98TGGGGASHH805 pKa = 7.74PEE807 pKa = 3.97AGDD810 pKa = 3.45NSSSDD815 pKa = 3.44RR816 pKa = 11.84NDD818 pKa = 3.92RR819 pKa = 11.84DD820 pKa = 4.52DD821 pKa = 6.38DD822 pKa = 3.69DD823 pKa = 4.39TEE825 pKa = 5.53NIDD828 pKa = 4.16EE829 pKa = 4.52EE830 pKa = 4.78DD831 pKa = 3.72VPLTEE836 pKa = 5.25GKK838 pKa = 10.3VADD841 pKa = 4.64FDD843 pKa = 5.64DD844 pKa = 4.63VPADD848 pKa = 3.01AWFAEE853 pKa = 4.28AVQYY857 pKa = 10.99VYY859 pKa = 10.98EE860 pKa = 4.92HH861 pKa = 7.26DD862 pKa = 4.22LMTGVSEE869 pKa = 5.3NLFAPNAQMNRR880 pKa = 11.84AMVAQILFNVEE891 pKa = 3.7KK892 pKa = 10.07PADD895 pKa = 3.95TEE897 pKa = 3.97APAAFRR903 pKa = 11.84DD904 pKa = 3.87VAPDD908 pKa = 2.71AWYY911 pKa = 10.76AEE913 pKa = 4.21AVNWAVWQGYY923 pKa = 6.42MSGYY927 pKa = 9.67GAGSFGPNDD936 pKa = 3.53ALTRR940 pKa = 11.84EE941 pKa = 4.14QLVTVLWRR949 pKa = 11.84YY950 pKa = 9.76SGSPVMGDD958 pKa = 2.69SSMLNTFSDD967 pKa = 3.82AALTSDD973 pKa = 3.92YY974 pKa = 10.87AQQAMIWAYY983 pKa = 9.61AQGVISGNADD993 pKa = 3.14GTLNPQGTATRR1004 pKa = 11.84AEE1006 pKa = 4.18IAQMLMNYY1014 pKa = 9.52CEE1016 pKa = 4.09NVKK1019 pKa = 10.63
MM1 pKa = 7.8KK2 pKa = 10.25KK3 pKa = 10.24RR4 pKa = 11.84FWACFLSLSLVLTMMPTMAFAADD27 pKa = 4.03DD28 pKa = 4.06EE29 pKa = 4.97VNEE32 pKa = 4.27PGGTVEE38 pKa = 4.21VCVEE42 pKa = 4.21TEE44 pKa = 3.95GCTLEE49 pKa = 4.75AGHH52 pKa = 6.51EE53 pKa = 4.55GEE55 pKa = 4.53CVVVEE60 pKa = 4.4SEE62 pKa = 4.26PEE64 pKa = 4.0EE65 pKa = 4.64PEE67 pKa = 4.05SAPCKK72 pKa = 8.55VTEE75 pKa = 4.32GCTLEE80 pKa = 4.35NGHH83 pKa = 6.37EE84 pKa = 4.54GEE86 pKa = 4.51CVVAEE91 pKa = 4.41SEE93 pKa = 4.33PEE95 pKa = 3.94EE96 pKa = 4.62PEE98 pKa = 4.47SVPCTVTEE106 pKa = 4.35GCTLEE111 pKa = 5.22DD112 pKa = 3.59GHH114 pKa = 7.05EE115 pKa = 4.57GGCVVEE121 pKa = 4.9SSVDD125 pKa = 3.51SLSEE129 pKa = 4.01DD130 pKa = 3.61EE131 pKa = 5.23EE132 pKa = 4.36IVISITEE139 pKa = 3.96ADD141 pKa = 3.78MNSADD146 pKa = 4.8EE147 pKa = 4.39EE148 pKa = 4.31NPPIKK153 pKa = 10.39SAIARR158 pKa = 11.84LSLADD163 pKa = 4.02PSSVTQLKK171 pKa = 10.08VVTEE175 pKa = 4.31GEE177 pKa = 4.53AYY179 pKa = 10.75LSPEE183 pKa = 4.0DD184 pKa = 3.52NEE186 pKa = 4.47YY187 pKa = 10.67IKK189 pKa = 10.0TVFSSLNYY197 pKa = 9.97LDD199 pKa = 5.45EE200 pKa = 4.95SEE202 pKa = 4.89CQCSTRR208 pKa = 11.84KK209 pKa = 8.44LTEE212 pKa = 4.31AYY214 pKa = 9.75LGKK217 pKa = 10.46SGLPDD222 pKa = 3.05EE223 pKa = 5.66LYY225 pKa = 11.15AEE227 pKa = 4.63FGGLSGYY234 pKa = 9.26TSLEE238 pKa = 3.87TLILPEE244 pKa = 3.91EE245 pKa = 4.34TQVVNSYY252 pKa = 8.46TLSEE256 pKa = 4.27RR257 pKa = 11.84GGGVNDD263 pKa = 3.86TGLTEE268 pKa = 5.56LNLSNNILVIEE279 pKa = 4.39SGAFSGNTNFVGNLVIPDD297 pKa = 3.59SVVFIGNNSFGVGNEE312 pKa = 4.73LIACGTLTLGDD323 pKa = 3.79SVKK326 pKa = 11.13YY327 pKa = 10.23IDD329 pKa = 4.39ANGFKK334 pKa = 10.57SRR336 pKa = 11.84LFEE339 pKa = 4.89GDD341 pKa = 3.75LVIPNSVEE349 pKa = 4.13MINNWSFSPGAFKK362 pKa = 11.05NGTWTLGDD370 pKa = 3.48NLINIGSAALKK381 pKa = 9.43GICSGNSGTFIVLNQYY397 pKa = 10.66KK398 pKa = 9.63MSTQAFADD406 pKa = 3.49NSFEE410 pKa = 4.42KK411 pKa = 10.45IAFEE415 pKa = 4.73EE416 pKa = 5.46GITDD420 pKa = 3.54ISTSQVIASSKK431 pKa = 9.94NIEE434 pKa = 4.36EE435 pKa = 4.08IVLPSTLEE443 pKa = 4.32SISGDD448 pKa = 3.71SAFAGCPALTTINFPKK464 pKa = 10.41SLEE467 pKa = 4.07RR468 pKa = 11.84LGDD471 pKa = 3.45RR472 pKa = 11.84TFEE475 pKa = 4.26NDD477 pKa = 3.29DD478 pKa = 3.66ALTSVVLPSNLTQIGSFTFEE498 pKa = 4.2DD499 pKa = 3.8SGILGLYY506 pKa = 9.52IPEE509 pKa = 4.27SVTDD513 pKa = 3.21IGNRR517 pKa = 11.84IVQDD521 pKa = 3.48IPSGSIVYY529 pKa = 8.81VANEE533 pKa = 3.65ALLNMMNEE541 pKa = 4.74SDD543 pKa = 3.91SAWNNKK549 pKa = 8.03YY550 pKa = 10.84NNEE553 pKa = 3.92TTALAMTNGGSFKK566 pKa = 10.67EE567 pKa = 3.99DD568 pKa = 3.37TEE570 pKa = 4.43FSEE573 pKa = 5.47SVLATPIKK581 pKa = 10.2SGNIFAGWYY590 pKa = 9.96ADD592 pKa = 3.87EE593 pKa = 4.49EE594 pKa = 4.37LADD597 pKa = 3.9EE598 pKa = 4.6VVTNADD604 pKa = 3.4AGNTYY609 pKa = 9.4YY610 pKa = 11.13AEE612 pKa = 4.19WLPSNYY618 pKa = 9.25TVTEE622 pKa = 4.25NINFGSVVYY631 pKa = 10.39GSTASRR637 pKa = 11.84VIQVNGSDD645 pKa = 3.29VAGALKK651 pKa = 10.77VAGNDD656 pKa = 3.09IFDD659 pKa = 3.95TVVNEE664 pKa = 4.43DD665 pKa = 3.1NTITVTPHH673 pKa = 6.06SNLSVGEE680 pKa = 4.13YY681 pKa = 10.88EE682 pKa = 3.82EE683 pKa = 4.27TLYY686 pKa = 9.85VTTPDD691 pKa = 3.09NATFFVPVTLEE702 pKa = 3.95VVPASSKK709 pKa = 11.17LEE711 pKa = 3.52IWANGAKK718 pKa = 10.07KK719 pKa = 10.44LSLSGGGNVTLTITGDD735 pKa = 3.6LNADD739 pKa = 3.76EE740 pKa = 4.67VTVTCNNEE748 pKa = 4.1NIEE751 pKa = 4.25VTKK754 pKa = 11.0NADD757 pKa = 3.17DD758 pKa = 4.36TYY760 pKa = 11.21TVTLPNEE767 pKa = 3.98TADD770 pKa = 3.64YY771 pKa = 9.53TFTATYY777 pKa = 10.67DD778 pKa = 3.31GDD780 pKa = 4.15QNHH783 pKa = 5.92EE784 pKa = 4.22MARR787 pKa = 11.84ATCTVSVTRR796 pKa = 11.84HH797 pKa = 4.98TGGGGASHH805 pKa = 7.74PEE807 pKa = 3.97AGDD810 pKa = 3.45NSSSDD815 pKa = 3.44RR816 pKa = 11.84NDD818 pKa = 3.92RR819 pKa = 11.84DD820 pKa = 4.52DD821 pKa = 6.38DD822 pKa = 3.69DD823 pKa = 4.39TEE825 pKa = 5.53NIDD828 pKa = 4.16EE829 pKa = 4.52EE830 pKa = 4.78DD831 pKa = 3.72VPLTEE836 pKa = 5.25GKK838 pKa = 10.3VADD841 pKa = 4.64FDD843 pKa = 5.64DD844 pKa = 4.63VPADD848 pKa = 3.01AWFAEE853 pKa = 4.28AVQYY857 pKa = 10.99VYY859 pKa = 10.98EE860 pKa = 4.92HH861 pKa = 7.26DD862 pKa = 4.22LMTGVSEE869 pKa = 5.3NLFAPNAQMNRR880 pKa = 11.84AMVAQILFNVEE891 pKa = 3.7KK892 pKa = 10.07PADD895 pKa = 3.95TEE897 pKa = 3.97APAAFRR903 pKa = 11.84DD904 pKa = 3.87VAPDD908 pKa = 2.71AWYY911 pKa = 10.76AEE913 pKa = 4.21AVNWAVWQGYY923 pKa = 6.42MSGYY927 pKa = 9.67GAGSFGPNDD936 pKa = 3.53ALTRR940 pKa = 11.84EE941 pKa = 4.14QLVTVLWRR949 pKa = 11.84YY950 pKa = 9.76SGSPVMGDD958 pKa = 2.69SSMLNTFSDD967 pKa = 3.82AALTSDD973 pKa = 3.92YY974 pKa = 10.87AQQAMIWAYY983 pKa = 9.61AQGVISGNADD993 pKa = 3.14GTLNPQGTATRR1004 pKa = 11.84AEE1006 pKa = 4.18IAQMLMNYY1014 pKa = 9.52CEE1016 pKa = 4.09NVKK1019 pKa = 10.63
Molecular weight: 109.17 kDa
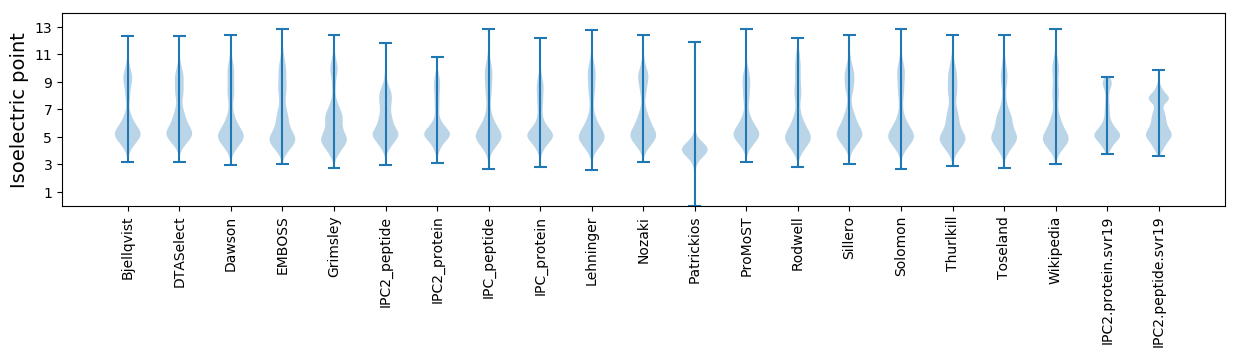
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R8VXU9|R8VXU9_9CLOT Uncharacterized protein OS=Butyricicoccus pullicaecorum 1.2 OX=1203606 GN=HMPREF1526_02727 PE=4 SV=1
MM1 pKa = 7.17VKK3 pKa = 10.19GVSKK7 pKa = 10.17RR8 pKa = 11.84VVVVRR13 pKa = 11.84PDD15 pKa = 3.2DD16 pKa = 3.69SRR18 pKa = 11.84FFEE21 pKa = 3.92QAFFIVRR28 pKa = 11.84EE29 pKa = 4.18GRR31 pKa = 11.84TASGDD36 pKa = 3.69ALRR39 pKa = 11.84EE40 pKa = 4.04ACALAARR47 pKa = 11.84YY48 pKa = 7.74HH49 pKa = 6.48AKK51 pKa = 10.37HH52 pKa = 6.51PARR55 pKa = 11.84PLPLRR60 pKa = 11.84RR61 pKa = 11.84YY62 pKa = 6.59TAPQMVLSALFGSGLMGAAWAVSCLVFF89 pKa = 5.28
MM1 pKa = 7.17VKK3 pKa = 10.19GVSKK7 pKa = 10.17RR8 pKa = 11.84VVVVRR13 pKa = 11.84PDD15 pKa = 3.2DD16 pKa = 3.69SRR18 pKa = 11.84FFEE21 pKa = 3.92QAFFIVRR28 pKa = 11.84EE29 pKa = 4.18GRR31 pKa = 11.84TASGDD36 pKa = 3.69ALRR39 pKa = 11.84EE40 pKa = 4.04ACALAARR47 pKa = 11.84YY48 pKa = 7.74HH49 pKa = 6.48AKK51 pKa = 10.37HH52 pKa = 6.51PARR55 pKa = 11.84PLPLRR60 pKa = 11.84RR61 pKa = 11.84YY62 pKa = 6.59TAPQMVLSALFGSGLMGAAWAVSCLVFF89 pKa = 5.28
Molecular weight: 9.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
955345 |
30 |
3162 |
307.5 |
34.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.491 ± 0.056 | 1.713 ± 0.02 |
5.93 ± 0.041 | 6.507 ± 0.052 |
3.78 ± 0.032 | 7.482 ± 0.041 |
2.012 ± 0.024 | 6.2 ± 0.041 |
4.831 ± 0.046 | 9.474 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.874 ± 0.027 | 3.429 ± 0.029 |
4.034 ± 0.025 | 3.876 ± 0.031 |
5.385 ± 0.047 | 5.52 ± 0.037 |
5.938 ± 0.042 | 7.034 ± 0.04 |
1.0 ± 0.014 | 3.49 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
