
Algibacter alginicilyticus
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Algibacter
Average proteome isoelectric point is 6.62
Get precalculated fractions of proteins
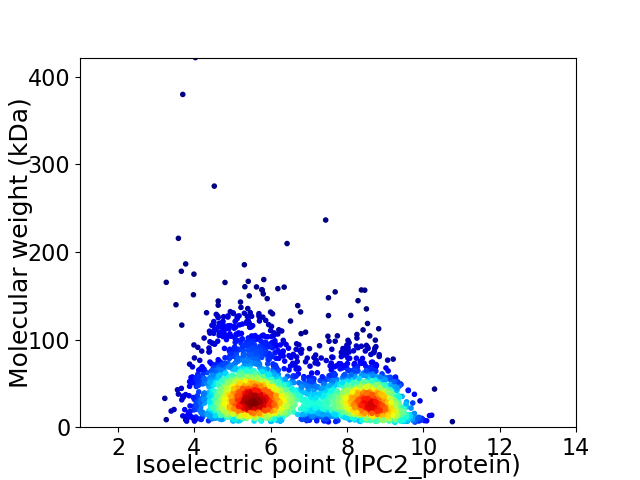
Virtual 2D-PAGE plot for 3236 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0P0DD31|A0A0P0DD31_9FLAO Altronate hydrolase OS=Algibacter alginicilyticus OX=1736674 GN=APS56_12635 PE=3 SV=1
MM1 pKa = 7.18ITKK4 pKa = 10.01QSLRR8 pKa = 11.84IFLSTVFFSFFIFLSCDD25 pKa = 3.39DD26 pKa = 4.72DD27 pKa = 5.85NEE29 pKa = 4.77INDD32 pKa = 3.63ATNPNFSNLITSISSAPGLEE52 pKa = 4.73FIFEE56 pKa = 4.29GTISDD61 pKa = 4.15DD62 pKa = 4.12NGIEE66 pKa = 4.23NININYY72 pKa = 9.23DD73 pKa = 2.64NWYY76 pKa = 8.56VDD78 pKa = 3.4KK79 pKa = 11.17NITFDD84 pKa = 4.03EE85 pKa = 4.31PLKK88 pKa = 10.65EE89 pKa = 3.91YY90 pKa = 10.72NLNYY94 pKa = 10.04KK95 pKa = 10.18FLVPEE100 pKa = 4.13EE101 pKa = 4.44EE102 pKa = 4.74EE103 pKa = 4.49PNSSHH108 pKa = 6.47TIKK111 pKa = 10.62ISATDD116 pKa = 3.17IAGNITTYY124 pKa = 11.21DD125 pKa = 3.63VIVSLDD131 pKa = 3.61YY132 pKa = 10.04DD133 pKa = 3.75TTLPQVAFISPISGSSNTVGDD154 pKa = 4.04LVEE157 pKa = 4.56LNIAFSDD164 pKa = 3.75NKK166 pKa = 10.19VLDD169 pKa = 4.14SIIVKK174 pKa = 9.14TEE176 pKa = 3.7SLDD179 pKa = 3.81YY180 pKa = 10.39EE181 pKa = 5.03VKK183 pKa = 10.85LKK185 pKa = 10.69MPDD188 pKa = 2.81NTLNYY193 pKa = 10.18NFTDD197 pKa = 3.46SVEE200 pKa = 4.1IPLTGISGAIEE211 pKa = 3.96FTATGIDD218 pKa = 3.33KK219 pKa = 10.14TGNTTTVYY227 pKa = 9.54STILVGEE234 pKa = 4.09KK235 pKa = 10.75DD236 pKa = 4.1EE237 pKa = 4.43IFNMYY242 pKa = 10.62AVGTSTWYY250 pKa = 10.07EE251 pKa = 3.82WDD253 pKa = 3.52PSKK256 pKa = 10.75ATEE259 pKa = 3.77MWKK262 pKa = 10.49NPDD265 pKa = 3.37NNDD268 pKa = 2.78WFVLEE273 pKa = 5.26FYY275 pKa = 10.08YY276 pKa = 11.09TMGNGVKK283 pKa = 10.49FIGQLDD289 pKa = 3.85WEE291 pKa = 4.63PNNWGTDD298 pKa = 3.59PNDD301 pKa = 3.06SSKK304 pKa = 10.7IINSQDD310 pKa = 2.79SGTIEE315 pKa = 4.19FPEE318 pKa = 3.95EE319 pKa = 4.1GYY321 pKa = 10.8YY322 pKa = 10.3HH323 pKa = 7.47VEE325 pKa = 3.93FNPYY329 pKa = 7.75TLEE332 pKa = 3.81YY333 pKa = 8.93TYY335 pKa = 11.31EE336 pKa = 4.15KK337 pKa = 10.43MEE339 pKa = 3.83VDD341 pKa = 3.32VDD343 pKa = 4.07VKK345 pKa = 11.35EE346 pKa = 4.14NMYY349 pKa = 11.46LMGNGFAGYY358 pKa = 10.53DD359 pKa = 4.0LDD361 pKa = 4.49WNPADD366 pKa = 5.25AIPMEE371 pKa = 5.0KK372 pKa = 10.26DD373 pKa = 3.41SNGNPYY379 pKa = 10.29VFTINVEE386 pKa = 3.96ITEE389 pKa = 4.14DD390 pKa = 3.36TSLKK394 pKa = 10.79FIGQTDD400 pKa = 3.21GWSPFDD406 pKa = 4.04CGFEE410 pKa = 4.26VGGEE414 pKa = 4.16TILPVNYY421 pKa = 9.45IKK423 pKa = 10.94CKK425 pKa = 10.18TGDD428 pKa = 3.26GSQDD432 pKa = 3.25LKK434 pKa = 11.21FKK436 pKa = 10.48NQAGTYY442 pKa = 8.5TITFDD447 pKa = 3.76YY448 pKa = 10.62FLLRR452 pKa = 11.84ATIHH456 pKa = 5.91QYY458 pKa = 10.0NN459 pKa = 3.49
MM1 pKa = 7.18ITKK4 pKa = 10.01QSLRR8 pKa = 11.84IFLSTVFFSFFIFLSCDD25 pKa = 3.39DD26 pKa = 4.72DD27 pKa = 5.85NEE29 pKa = 4.77INDD32 pKa = 3.63ATNPNFSNLITSISSAPGLEE52 pKa = 4.73FIFEE56 pKa = 4.29GTISDD61 pKa = 4.15DD62 pKa = 4.12NGIEE66 pKa = 4.23NININYY72 pKa = 9.23DD73 pKa = 2.64NWYY76 pKa = 8.56VDD78 pKa = 3.4KK79 pKa = 11.17NITFDD84 pKa = 4.03EE85 pKa = 4.31PLKK88 pKa = 10.65EE89 pKa = 3.91YY90 pKa = 10.72NLNYY94 pKa = 10.04KK95 pKa = 10.18FLVPEE100 pKa = 4.13EE101 pKa = 4.44EE102 pKa = 4.74EE103 pKa = 4.49PNSSHH108 pKa = 6.47TIKK111 pKa = 10.62ISATDD116 pKa = 3.17IAGNITTYY124 pKa = 11.21DD125 pKa = 3.63VIVSLDD131 pKa = 3.61YY132 pKa = 10.04DD133 pKa = 3.75TTLPQVAFISPISGSSNTVGDD154 pKa = 4.04LVEE157 pKa = 4.56LNIAFSDD164 pKa = 3.75NKK166 pKa = 10.19VLDD169 pKa = 4.14SIIVKK174 pKa = 9.14TEE176 pKa = 3.7SLDD179 pKa = 3.81YY180 pKa = 10.39EE181 pKa = 5.03VKK183 pKa = 10.85LKK185 pKa = 10.69MPDD188 pKa = 2.81NTLNYY193 pKa = 10.18NFTDD197 pKa = 3.46SVEE200 pKa = 4.1IPLTGISGAIEE211 pKa = 3.96FTATGIDD218 pKa = 3.33KK219 pKa = 10.14TGNTTTVYY227 pKa = 9.54STILVGEE234 pKa = 4.09KK235 pKa = 10.75DD236 pKa = 4.1EE237 pKa = 4.43IFNMYY242 pKa = 10.62AVGTSTWYY250 pKa = 10.07EE251 pKa = 3.82WDD253 pKa = 3.52PSKK256 pKa = 10.75ATEE259 pKa = 3.77MWKK262 pKa = 10.49NPDD265 pKa = 3.37NNDD268 pKa = 2.78WFVLEE273 pKa = 5.26FYY275 pKa = 10.08YY276 pKa = 11.09TMGNGVKK283 pKa = 10.49FIGQLDD289 pKa = 3.85WEE291 pKa = 4.63PNNWGTDD298 pKa = 3.59PNDD301 pKa = 3.06SSKK304 pKa = 10.7IINSQDD310 pKa = 2.79SGTIEE315 pKa = 4.19FPEE318 pKa = 3.95EE319 pKa = 4.1GYY321 pKa = 10.8YY322 pKa = 10.3HH323 pKa = 7.47VEE325 pKa = 3.93FNPYY329 pKa = 7.75TLEE332 pKa = 3.81YY333 pKa = 8.93TYY335 pKa = 11.31EE336 pKa = 4.15KK337 pKa = 10.43MEE339 pKa = 3.83VDD341 pKa = 3.32VDD343 pKa = 4.07VKK345 pKa = 11.35EE346 pKa = 4.14NMYY349 pKa = 11.46LMGNGFAGYY358 pKa = 10.53DD359 pKa = 4.0LDD361 pKa = 4.49WNPADD366 pKa = 5.25AIPMEE371 pKa = 5.0KK372 pKa = 10.26DD373 pKa = 3.41SNGNPYY379 pKa = 10.29VFTINVEE386 pKa = 3.96ITEE389 pKa = 4.14DD390 pKa = 3.36TSLKK394 pKa = 10.79FIGQTDD400 pKa = 3.21GWSPFDD406 pKa = 4.04CGFEE410 pKa = 4.26VGGEE414 pKa = 4.16TILPVNYY421 pKa = 9.45IKK423 pKa = 10.94CKK425 pKa = 10.18TGDD428 pKa = 3.26GSQDD432 pKa = 3.25LKK434 pKa = 11.21FKK436 pKa = 10.48NQAGTYY442 pKa = 8.5TITFDD447 pKa = 3.76YY448 pKa = 10.62FLLRR452 pKa = 11.84ATIHH456 pKa = 5.91QYY458 pKa = 10.0NN459 pKa = 3.49
Molecular weight: 51.91 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0P0CQ50|A0A0P0CQ50_9FLAO Uncharacterized protein OS=Algibacter alginicilyticus OX=1736674 GN=APS56_07705 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 7.97KK42 pKa = 10.62LSVSSEE48 pKa = 4.03TRR50 pKa = 11.84HH51 pKa = 5.96KK52 pKa = 10.68KK53 pKa = 9.8
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 7.97KK42 pKa = 10.62LSVSSEE48 pKa = 4.03TRR50 pKa = 11.84HH51 pKa = 5.96KK52 pKa = 10.68KK53 pKa = 9.8
Molecular weight: 6.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1177855 |
50 |
3879 |
364.0 |
41.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.972 ± 0.04 | 0.735 ± 0.014 |
5.593 ± 0.03 | 6.346 ± 0.035 |
5.223 ± 0.033 | 6.243 ± 0.045 |
1.81 ± 0.021 | 8.363 ± 0.045 |
7.953 ± 0.055 | 9.152 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.065 ± 0.022 | 6.886 ± 0.043 |
3.328 ± 0.022 | 3.243 ± 0.024 |
3.137 ± 0.025 | 6.641 ± 0.035 |
5.955 ± 0.043 | 6.025 ± 0.032 |
1.08 ± 0.016 | 4.249 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
