
BK polyomavirus (BKPyV) (Human polyomavirus 1)
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Betapolyomavirus
Average proteome isoelectric point is 7.1
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

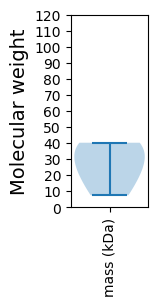
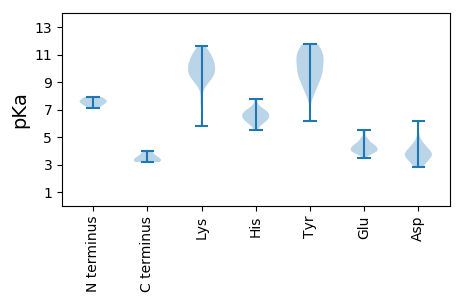
Protein with the lowest isoelectric point:
>tr|Q0PCM6|Q0PCM6_POVBK Minor capsid protein OS=BK polyomavirus OX=1891762 PE=3 SV=1
MM1 pKa = 7.71APTKK5 pKa = 10.54RR6 pKa = 11.84KK7 pKa = 10.25GEE9 pKa = 4.26CPGAAPKK16 pKa = 10.27KK17 pKa = 9.07PKK19 pKa = 10.55EE20 pKa = 3.96PVQVPKK26 pKa = 10.86LLIKK30 pKa = 10.81GGVEE34 pKa = 3.79VLEE37 pKa = 4.53VKK39 pKa = 9.99TGVDD43 pKa = 3.63AITEE47 pKa = 4.29VEE49 pKa = 4.37CFLNPEE55 pKa = 4.38MGDD58 pKa = 3.29PDD60 pKa = 3.91NDD62 pKa = 3.24LRR64 pKa = 11.84GYY66 pKa = 8.45SLRR69 pKa = 11.84LTAEE73 pKa = 3.95TAFDD77 pKa = 3.71SDD79 pKa = 3.96SPDD82 pKa = 3.09RR83 pKa = 11.84KK84 pKa = 9.47MLPCYY89 pKa = 9.18STARR93 pKa = 11.84IPLPNLNEE101 pKa = 4.72DD102 pKa = 3.98LTCGNLLMWEE112 pKa = 4.08AVTVKK117 pKa = 10.14TEE119 pKa = 4.19VIGITSMLNLHH130 pKa = 6.71AGSQKK135 pKa = 8.81VHH137 pKa = 6.45EE138 pKa = 4.91NGGGKK143 pKa = 9.33PIQGSNFHH151 pKa = 6.63FFAVGGDD158 pKa = 3.83PLEE161 pKa = 4.33MQGVLMNYY169 pKa = 6.15RR170 pKa = 11.84TKK172 pKa = 10.72YY173 pKa = 10.14PEE175 pKa = 4.09GTVTPKK181 pKa = 10.87NPTAQSQVMNTDD193 pKa = 2.82HH194 pKa = 7.79KK195 pKa = 11.53AYY197 pKa = 10.42LDD199 pKa = 3.64KK200 pKa = 11.64NNAYY204 pKa = 9.1PVEE207 pKa = 4.23CWIPDD212 pKa = 3.47PSRR215 pKa = 11.84NEE217 pKa = 3.45NTRR220 pKa = 11.84YY221 pKa = 9.81FGTYY225 pKa = 8.7TGGEE229 pKa = 4.01NVPPVLHH236 pKa = 5.52VTNTATTVLLDD247 pKa = 3.84EE248 pKa = 4.66QGVGPLCKK256 pKa = 9.76ADD258 pKa = 3.67SLYY261 pKa = 11.29VSAADD266 pKa = 3.09ICGLFTNSSGTQQWRR281 pKa = 11.84GLPRR285 pKa = 11.84YY286 pKa = 9.33FKK288 pKa = 10.22IRR290 pKa = 11.84LRR292 pKa = 11.84KK293 pKa = 9.4RR294 pKa = 11.84SVKK297 pKa = 10.18NPYY300 pKa = 9.28PISFLLSDD308 pKa = 4.93LINRR312 pKa = 11.84RR313 pKa = 11.84TQRR316 pKa = 11.84VDD318 pKa = 3.18GQPMYY323 pKa = 11.3GMEE326 pKa = 4.2SQVEE330 pKa = 4.21EE331 pKa = 4.01VRR333 pKa = 11.84VFDD336 pKa = 4.45GTEE339 pKa = 3.84QLPGDD344 pKa = 4.33PDD346 pKa = 3.84MIRR349 pKa = 11.84YY350 pKa = 8.96IDD352 pKa = 3.87RR353 pKa = 11.84QGQLQTKK360 pKa = 7.5MVV362 pKa = 3.43
MM1 pKa = 7.71APTKK5 pKa = 10.54RR6 pKa = 11.84KK7 pKa = 10.25GEE9 pKa = 4.26CPGAAPKK16 pKa = 10.27KK17 pKa = 9.07PKK19 pKa = 10.55EE20 pKa = 3.96PVQVPKK26 pKa = 10.86LLIKK30 pKa = 10.81GGVEE34 pKa = 3.79VLEE37 pKa = 4.53VKK39 pKa = 9.99TGVDD43 pKa = 3.63AITEE47 pKa = 4.29VEE49 pKa = 4.37CFLNPEE55 pKa = 4.38MGDD58 pKa = 3.29PDD60 pKa = 3.91NDD62 pKa = 3.24LRR64 pKa = 11.84GYY66 pKa = 8.45SLRR69 pKa = 11.84LTAEE73 pKa = 3.95TAFDD77 pKa = 3.71SDD79 pKa = 3.96SPDD82 pKa = 3.09RR83 pKa = 11.84KK84 pKa = 9.47MLPCYY89 pKa = 9.18STARR93 pKa = 11.84IPLPNLNEE101 pKa = 4.72DD102 pKa = 3.98LTCGNLLMWEE112 pKa = 4.08AVTVKK117 pKa = 10.14TEE119 pKa = 4.19VIGITSMLNLHH130 pKa = 6.71AGSQKK135 pKa = 8.81VHH137 pKa = 6.45EE138 pKa = 4.91NGGGKK143 pKa = 9.33PIQGSNFHH151 pKa = 6.63FFAVGGDD158 pKa = 3.83PLEE161 pKa = 4.33MQGVLMNYY169 pKa = 6.15RR170 pKa = 11.84TKK172 pKa = 10.72YY173 pKa = 10.14PEE175 pKa = 4.09GTVTPKK181 pKa = 10.87NPTAQSQVMNTDD193 pKa = 2.82HH194 pKa = 7.79KK195 pKa = 11.53AYY197 pKa = 10.42LDD199 pKa = 3.64KK200 pKa = 11.64NNAYY204 pKa = 9.1PVEE207 pKa = 4.23CWIPDD212 pKa = 3.47PSRR215 pKa = 11.84NEE217 pKa = 3.45NTRR220 pKa = 11.84YY221 pKa = 9.81FGTYY225 pKa = 8.7TGGEE229 pKa = 4.01NVPPVLHH236 pKa = 5.52VTNTATTVLLDD247 pKa = 3.84EE248 pKa = 4.66QGVGPLCKK256 pKa = 9.76ADD258 pKa = 3.67SLYY261 pKa = 11.29VSAADD266 pKa = 3.09ICGLFTNSSGTQQWRR281 pKa = 11.84GLPRR285 pKa = 11.84YY286 pKa = 9.33FKK288 pKa = 10.22IRR290 pKa = 11.84LRR292 pKa = 11.84KK293 pKa = 9.4RR294 pKa = 11.84SVKK297 pKa = 10.18NPYY300 pKa = 9.28PISFLLSDD308 pKa = 4.93LINRR312 pKa = 11.84RR313 pKa = 11.84TQRR316 pKa = 11.84VDD318 pKa = 3.18GQPMYY323 pKa = 11.3GMEE326 pKa = 4.2SQVEE330 pKa = 4.21EE331 pKa = 4.01VRR333 pKa = 11.84VFDD336 pKa = 4.45GTEE339 pKa = 3.84QLPGDD344 pKa = 4.33PDD346 pKa = 3.84MIRR349 pKa = 11.84YY350 pKa = 8.96IDD352 pKa = 3.87RR353 pKa = 11.84QGQLQTKK360 pKa = 7.5MVV362 pKa = 3.43
Molecular weight: 40.16 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q3C215|Q3C215_POVBK Small t antigen OS=BK polyomavirus OX=1891762 PE=4 SV=1
MM1 pKa = 7.53VLRR4 pKa = 11.84QLSRR8 pKa = 11.84QASVKK13 pKa = 10.14VSKK16 pKa = 8.91TWTGTKK22 pKa = 9.78KK23 pKa = 9.72RR24 pKa = 11.84AQRR27 pKa = 11.84ILIFILEE34 pKa = 4.41LLLEE38 pKa = 4.51FCRR41 pKa = 11.84GEE43 pKa = 4.32DD44 pKa = 4.5SVDD47 pKa = 3.39GKK49 pKa = 10.79NKK51 pKa = 9.27STTALPAVKK60 pKa = 10.32DD61 pKa = 3.9SVKK64 pKa = 10.92DD65 pKa = 3.45SS66 pKa = 3.5
MM1 pKa = 7.53VLRR4 pKa = 11.84QLSRR8 pKa = 11.84QASVKK13 pKa = 10.14VSKK16 pKa = 8.91TWTGTKK22 pKa = 9.78KK23 pKa = 9.72RR24 pKa = 11.84AQRR27 pKa = 11.84ILIFILEE34 pKa = 4.41LLLEE38 pKa = 4.51FCRR41 pKa = 11.84GEE43 pKa = 4.32DD44 pKa = 4.5SVDD47 pKa = 3.39GKK49 pKa = 10.79NKK51 pKa = 9.27STTALPAVKK60 pKa = 10.32DD61 pKa = 3.9SVKK64 pKa = 10.92DD65 pKa = 3.45SS66 pKa = 3.5
Molecular weight: 7.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1183 |
66 |
362 |
236.6 |
26.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.27 ± 2.022 | 1.606 ± 1.086 |
5.325 ± 0.624 | 6.255 ± 0.291 |
3.635 ± 0.294 | 6.593 ± 0.805 |
1.522 ± 0.191 | 4.818 ± 0.752 |
5.241 ± 1.292 | 8.791 ± 0.972 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.536 ± 0.596 | 4.48 ± 0.709 |
5.748 ± 0.914 | 4.818 ± 0.356 |
6.678 ± 0.601 | 6.34 ± 0.959 |
6.932 ± 0.597 | 6.34 ± 0.618 |
1.691 ± 0.53 | 3.381 ± 0.401 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
