
Labrenzia alba
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Stappiaceae; Labrenzia
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
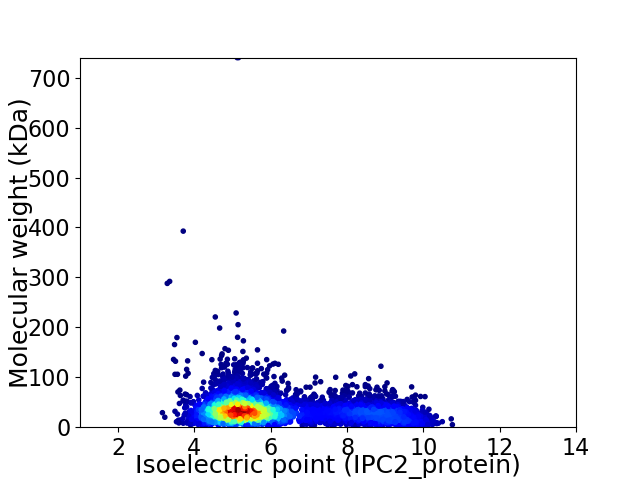
Virtual 2D-PAGE plot for 6265 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0M6ZV23|A0A0M6ZV23_9RHOB Bacterial regulatory proteins tetR family OS=Labrenzia alba OX=311410 GN=LA5096_00961 PE=4 SV=1
MM1 pKa = 7.52TKK3 pKa = 10.35PSFDD7 pKa = 3.74PAFLSVDD14 pKa = 3.16EE15 pKa = 4.69TMPDD19 pKa = 3.43PSTSTEE25 pKa = 3.83SHH27 pKa = 6.86AEE29 pKa = 3.96NLDD32 pKa = 3.64QASLPFFSLDD42 pKa = 3.68EE43 pKa = 4.43IADD46 pKa = 3.92QLTDD50 pKa = 4.41GYY52 pKa = 9.61WLNEE56 pKa = 3.73GGLRR60 pKa = 11.84RR61 pKa = 11.84AFDD64 pKa = 3.73TSSSNQVSVNIDD76 pKa = 3.38GLSEE80 pKa = 4.18EE81 pKa = 4.65GQSLAQRR88 pKa = 11.84ALTEE92 pKa = 4.08WSNVADD98 pKa = 3.97LEE100 pKa = 4.32FVYY103 pKa = 11.05VSGAAQITFNDD114 pKa = 3.63EE115 pKa = 3.7NTNASATSQTSGGMILSSEE134 pKa = 4.28VNIPLSYY141 pKa = 11.11LEE143 pKa = 4.02THH145 pKa = 5.92GTGVPGFAYY154 pKa = 10.29FMYY157 pKa = 10.39LRR159 pKa = 11.84EE160 pKa = 4.27IGQALGLGHH169 pKa = 7.49AGNYY173 pKa = 8.67EE174 pKa = 3.81DD175 pKa = 3.87TATYY179 pKa = 10.46GVDD182 pKa = 2.75NHH184 pKa = 6.22YY185 pKa = 11.57ANDD188 pKa = 3.42SWQSSVMSAFSQTDD202 pKa = 3.2NTSIDD207 pKa = 3.01ASFAYY212 pKa = 9.4PVTPMIADD220 pKa = 3.84ILAVQNLYY228 pKa = 11.09GEE230 pKa = 4.58PDD232 pKa = 4.08YY233 pKa = 10.9IYY235 pKa = 9.9PDD237 pKa = 3.5YY238 pKa = 10.91RR239 pKa = 11.84IFGANVYY246 pKa = 10.7GDD248 pKa = 3.53YY249 pKa = 10.94NEE251 pKa = 4.42RR252 pKa = 11.84GFYY255 pKa = 10.16EE256 pKa = 4.19ISPDD260 pKa = 3.11ATATIFDD267 pKa = 4.33SGGEE271 pKa = 3.87DD272 pKa = 4.22LIRR275 pKa = 11.84FLGYY279 pKa = 10.53SGDD282 pKa = 3.62QRR284 pKa = 11.84IDD286 pKa = 3.5LNQEE290 pKa = 3.61ATSDD294 pKa = 3.75FAGGKK299 pKa = 9.52GNLVIARR306 pKa = 11.84GAVIEE311 pKa = 4.17RR312 pKa = 11.84AYY314 pKa = 10.56GGNGNDD320 pKa = 3.68EE321 pKa = 4.77IIGNSSDD328 pKa = 3.7NLLVGYY334 pKa = 9.97DD335 pKa = 4.03GNDD338 pKa = 3.28TLIGGEE344 pKa = 4.21GDD346 pKa = 4.25DD347 pKa = 6.22FLSGWNGVDD356 pKa = 4.81LMFGGPGDD364 pKa = 4.07DD365 pKa = 4.01TIMATNEE372 pKa = 3.76DD373 pKa = 4.28VFFKK377 pKa = 10.86DD378 pKa = 4.06GAPIDD383 pKa = 4.14LGGPGYY389 pKa = 10.73DD390 pKa = 2.97RR391 pKa = 11.84LIISLEE397 pKa = 4.36SYY399 pKa = 10.98FEE401 pKa = 4.16TDD403 pKa = 2.76SLSQFGIEE411 pKa = 4.47HH412 pKa = 6.54FWGGGGDD419 pKa = 3.8DD420 pKa = 4.45RR421 pKa = 11.84AIGDD425 pKa = 4.88DD426 pKa = 3.7GTVDD430 pKa = 3.67YY431 pKa = 10.58ILEE434 pKa = 4.16GRR436 pKa = 11.84YY437 pKa = 10.65GNDD440 pKa = 2.58ILTGNAGNDD449 pKa = 3.48TLLGGDD455 pKa = 3.58GHH457 pKa = 7.28DD458 pKa = 4.1VLTGGAGNDD467 pKa = 3.75FLRR470 pKa = 11.84GGIWGYY476 pKa = 10.86DD477 pKa = 3.08QLFAQEE483 pKa = 4.83GDD485 pKa = 3.5DD486 pKa = 3.56TVYY489 pKa = 10.59YY490 pKa = 11.0ASGKK494 pKa = 8.75VFWDD498 pKa = 3.16DD499 pKa = 3.82HH500 pKa = 6.25KK501 pKa = 11.17PRR503 pKa = 11.84DD504 pKa = 3.56IGGSGFDD511 pKa = 3.28TLMIEE516 pKa = 4.47NGSKK520 pKa = 10.65FNTVGLSWYY529 pKa = 10.1GFEE532 pKa = 4.41AFEE535 pKa = 4.6GAEE538 pKa = 4.08KK539 pKa = 10.02TDD541 pKa = 3.09RR542 pKa = 11.84VYY544 pKa = 11.52GNDD547 pKa = 2.92ASVNYY552 pKa = 10.38RR553 pKa = 11.84MDD555 pKa = 3.78GGGGDD560 pKa = 3.92DD561 pKa = 4.82DD562 pKa = 5.34LRR564 pKa = 11.84GSGGNDD570 pKa = 2.89TLLGGEE576 pKa = 4.33GTDD579 pKa = 3.61ILFGKK584 pKa = 10.4DD585 pKa = 3.49GDD587 pKa = 4.63DD588 pKa = 4.53LLDD591 pKa = 4.92PGPSTSGQQEE601 pKa = 4.31LFGGDD606 pKa = 3.02GSDD609 pKa = 3.58TYY611 pKa = 10.94RR612 pKa = 11.84ISTDD616 pKa = 2.55SGRR619 pKa = 11.84TYY621 pKa = 10.35IEE623 pKa = 3.85EE624 pKa = 4.21LFEE627 pKa = 4.15QGSDD631 pKa = 3.41SLVFEE636 pKa = 4.87SLMPSDD642 pKa = 3.99IEE644 pKa = 4.08ATIVDD649 pKa = 4.17GDD651 pKa = 3.89MLLLSWHH658 pKa = 7.39DD659 pKa = 3.39GTGQVTVNDD668 pKa = 4.35LGQNIEE674 pKa = 4.18RR675 pKa = 11.84YY676 pKa = 9.35EE677 pKa = 4.04FADD680 pKa = 4.25GSAYY684 pKa = 10.7SADD687 pKa = 3.26EE688 pKa = 4.45FEE690 pKa = 5.06FSS692 pKa = 3.85
MM1 pKa = 7.52TKK3 pKa = 10.35PSFDD7 pKa = 3.74PAFLSVDD14 pKa = 3.16EE15 pKa = 4.69TMPDD19 pKa = 3.43PSTSTEE25 pKa = 3.83SHH27 pKa = 6.86AEE29 pKa = 3.96NLDD32 pKa = 3.64QASLPFFSLDD42 pKa = 3.68EE43 pKa = 4.43IADD46 pKa = 3.92QLTDD50 pKa = 4.41GYY52 pKa = 9.61WLNEE56 pKa = 3.73GGLRR60 pKa = 11.84RR61 pKa = 11.84AFDD64 pKa = 3.73TSSSNQVSVNIDD76 pKa = 3.38GLSEE80 pKa = 4.18EE81 pKa = 4.65GQSLAQRR88 pKa = 11.84ALTEE92 pKa = 4.08WSNVADD98 pKa = 3.97LEE100 pKa = 4.32FVYY103 pKa = 11.05VSGAAQITFNDD114 pKa = 3.63EE115 pKa = 3.7NTNASATSQTSGGMILSSEE134 pKa = 4.28VNIPLSYY141 pKa = 11.11LEE143 pKa = 4.02THH145 pKa = 5.92GTGVPGFAYY154 pKa = 10.29FMYY157 pKa = 10.39LRR159 pKa = 11.84EE160 pKa = 4.27IGQALGLGHH169 pKa = 7.49AGNYY173 pKa = 8.67EE174 pKa = 3.81DD175 pKa = 3.87TATYY179 pKa = 10.46GVDD182 pKa = 2.75NHH184 pKa = 6.22YY185 pKa = 11.57ANDD188 pKa = 3.42SWQSSVMSAFSQTDD202 pKa = 3.2NTSIDD207 pKa = 3.01ASFAYY212 pKa = 9.4PVTPMIADD220 pKa = 3.84ILAVQNLYY228 pKa = 11.09GEE230 pKa = 4.58PDD232 pKa = 4.08YY233 pKa = 10.9IYY235 pKa = 9.9PDD237 pKa = 3.5YY238 pKa = 10.91RR239 pKa = 11.84IFGANVYY246 pKa = 10.7GDD248 pKa = 3.53YY249 pKa = 10.94NEE251 pKa = 4.42RR252 pKa = 11.84GFYY255 pKa = 10.16EE256 pKa = 4.19ISPDD260 pKa = 3.11ATATIFDD267 pKa = 4.33SGGEE271 pKa = 3.87DD272 pKa = 4.22LIRR275 pKa = 11.84FLGYY279 pKa = 10.53SGDD282 pKa = 3.62QRR284 pKa = 11.84IDD286 pKa = 3.5LNQEE290 pKa = 3.61ATSDD294 pKa = 3.75FAGGKK299 pKa = 9.52GNLVIARR306 pKa = 11.84GAVIEE311 pKa = 4.17RR312 pKa = 11.84AYY314 pKa = 10.56GGNGNDD320 pKa = 3.68EE321 pKa = 4.77IIGNSSDD328 pKa = 3.7NLLVGYY334 pKa = 9.97DD335 pKa = 4.03GNDD338 pKa = 3.28TLIGGEE344 pKa = 4.21GDD346 pKa = 4.25DD347 pKa = 6.22FLSGWNGVDD356 pKa = 4.81LMFGGPGDD364 pKa = 4.07DD365 pKa = 4.01TIMATNEE372 pKa = 3.76DD373 pKa = 4.28VFFKK377 pKa = 10.86DD378 pKa = 4.06GAPIDD383 pKa = 4.14LGGPGYY389 pKa = 10.73DD390 pKa = 2.97RR391 pKa = 11.84LIISLEE397 pKa = 4.36SYY399 pKa = 10.98FEE401 pKa = 4.16TDD403 pKa = 2.76SLSQFGIEE411 pKa = 4.47HH412 pKa = 6.54FWGGGGDD419 pKa = 3.8DD420 pKa = 4.45RR421 pKa = 11.84AIGDD425 pKa = 4.88DD426 pKa = 3.7GTVDD430 pKa = 3.67YY431 pKa = 10.58ILEE434 pKa = 4.16GRR436 pKa = 11.84YY437 pKa = 10.65GNDD440 pKa = 2.58ILTGNAGNDD449 pKa = 3.48TLLGGDD455 pKa = 3.58GHH457 pKa = 7.28DD458 pKa = 4.1VLTGGAGNDD467 pKa = 3.75FLRR470 pKa = 11.84GGIWGYY476 pKa = 10.86DD477 pKa = 3.08QLFAQEE483 pKa = 4.83GDD485 pKa = 3.5DD486 pKa = 3.56TVYY489 pKa = 10.59YY490 pKa = 11.0ASGKK494 pKa = 8.75VFWDD498 pKa = 3.16DD499 pKa = 3.82HH500 pKa = 6.25KK501 pKa = 11.17PRR503 pKa = 11.84DD504 pKa = 3.56IGGSGFDD511 pKa = 3.28TLMIEE516 pKa = 4.47NGSKK520 pKa = 10.65FNTVGLSWYY529 pKa = 10.1GFEE532 pKa = 4.41AFEE535 pKa = 4.6GAEE538 pKa = 4.08KK539 pKa = 10.02TDD541 pKa = 3.09RR542 pKa = 11.84VYY544 pKa = 11.52GNDD547 pKa = 2.92ASVNYY552 pKa = 10.38RR553 pKa = 11.84MDD555 pKa = 3.78GGGGDD560 pKa = 3.92DD561 pKa = 4.82DD562 pKa = 5.34LRR564 pKa = 11.84GSGGNDD570 pKa = 2.89TLLGGEE576 pKa = 4.33GTDD579 pKa = 3.61ILFGKK584 pKa = 10.4DD585 pKa = 3.49GDD587 pKa = 4.63DD588 pKa = 4.53LLDD591 pKa = 4.92PGPSTSGQQEE601 pKa = 4.31LFGGDD606 pKa = 3.02GSDD609 pKa = 3.58TYY611 pKa = 10.94RR612 pKa = 11.84ISTDD616 pKa = 2.55SGRR619 pKa = 11.84TYY621 pKa = 10.35IEE623 pKa = 3.85EE624 pKa = 4.21LFEE627 pKa = 4.15QGSDD631 pKa = 3.41SLVFEE636 pKa = 4.87SLMPSDD642 pKa = 3.99IEE644 pKa = 4.08ATIVDD649 pKa = 4.17GDD651 pKa = 3.89MLLLSWHH658 pKa = 7.39DD659 pKa = 3.39GTGQVTVNDD668 pKa = 4.35LGQNIEE674 pKa = 4.18RR675 pKa = 11.84YY676 pKa = 9.35EE677 pKa = 4.04FADD680 pKa = 4.25GSAYY684 pKa = 10.7SADD687 pKa = 3.26EE688 pKa = 4.45FEE690 pKa = 5.06FSS692 pKa = 3.85
Molecular weight: 74.32 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0M6Z5K9|A0A0M6Z5K9_9RHOB L-threonine aldolase OS=Labrenzia alba OX=311410 GN=ltaE PE=3 SV=1
MM1 pKa = 7.68KK2 pKa = 10.17IKK4 pKa = 10.8NSLKK8 pKa = 10.5ALMNRR13 pKa = 11.84HH14 pKa = 6.1RR15 pKa = 11.84DD16 pKa = 3.08NRR18 pKa = 11.84MVRR21 pKa = 11.84RR22 pKa = 11.84KK23 pKa = 10.31GRR25 pKa = 11.84VFIINKK31 pKa = 8.97KK32 pKa = 8.7NPRR35 pKa = 11.84FKK37 pKa = 10.85ARR39 pKa = 11.84QGG41 pKa = 3.28
MM1 pKa = 7.68KK2 pKa = 10.17IKK4 pKa = 10.8NSLKK8 pKa = 10.5ALMNRR13 pKa = 11.84HH14 pKa = 6.1RR15 pKa = 11.84DD16 pKa = 3.08NRR18 pKa = 11.84MVRR21 pKa = 11.84RR22 pKa = 11.84KK23 pKa = 10.31GRR25 pKa = 11.84VFIINKK31 pKa = 8.97KK32 pKa = 8.7NPRR35 pKa = 11.84FKK37 pKa = 10.85ARR39 pKa = 11.84QGG41 pKa = 3.28
Molecular weight: 5.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2028616 |
29 |
6892 |
323.8 |
35.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.755 ± 0.038 | 0.946 ± 0.01 |
5.994 ± 0.03 | 6.148 ± 0.032 |
4.132 ± 0.025 | 8.331 ± 0.033 |
2.018 ± 0.015 | 5.508 ± 0.02 |
3.746 ± 0.021 | 10.143 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.514 ± 0.016 | 3.113 ± 0.019 |
4.773 ± 0.023 | 3.21 ± 0.017 |
6.16 ± 0.035 | 6.075 ± 0.024 |
5.444 ± 0.02 | 7.362 ± 0.022 |
1.321 ± 0.014 | 2.307 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
