
Collinsella intestinalis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria;
Average proteome isoelectric point is 5.69
Get precalculated fractions of proteins
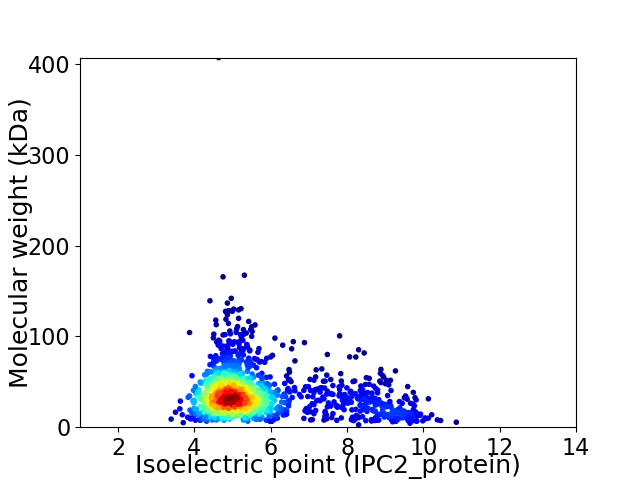
Virtual 2D-PAGE plot for 1430 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A414FU40|A0A414FU40_9ACTN ArsC family transcriptional regulator OS=Collinsella intestinalis OX=147207 GN=spxA PE=3 SV=1
MM1 pKa = 7.24MFLAGCGPAPQGSGDD16 pKa = 3.86AADD19 pKa = 4.81GSASASGILLGDD31 pKa = 4.09GKK33 pKa = 7.94TLRR36 pKa = 11.84VGMEE40 pKa = 3.77AAYY43 pKa = 10.13APYY46 pKa = 8.99NWQVSEE52 pKa = 4.13EE53 pKa = 4.44SEE55 pKa = 4.02FTIPIEE61 pKa = 4.2NVPGAYY67 pKa = 9.89ADD69 pKa = 4.36GYY71 pKa = 10.23DD72 pKa = 3.51VQVAKK77 pKa = 10.6IIADD81 pKa = 3.79GLGMEE86 pKa = 4.56PVAVKK91 pKa = 10.9LEE93 pKa = 4.07FGSLIDD99 pKa = 4.24ALNNGQVDD107 pKa = 3.91IVCAGMSVTPEE118 pKa = 3.58RR119 pKa = 11.84AAAAAFSDD127 pKa = 4.49SYY129 pKa = 11.82VDD131 pKa = 4.55DD132 pKa = 6.16DD133 pKa = 3.91IVMICRR139 pKa = 11.84ADD141 pKa = 3.26SAYY144 pKa = 10.71AGATTFDD151 pKa = 3.87EE152 pKa = 4.71LADD155 pKa = 3.96ASILGQAATMYY166 pKa = 10.76DD167 pKa = 3.67DD168 pKa = 5.47VIAQIEE174 pKa = 4.79GANHH178 pKa = 4.7MTPAEE183 pKa = 4.14TQPMVVEE190 pKa = 4.02NLRR193 pKa = 11.84NGTCDD198 pKa = 3.01IITYY202 pKa = 11.01SMLSAPKK209 pKa = 10.1LLEE212 pKa = 4.09VYY214 pKa = 9.98PDD216 pKa = 3.72LVVLDD221 pKa = 4.45MEE223 pKa = 4.95DD224 pKa = 3.42AFEE227 pKa = 4.72GSLMPDD233 pKa = 2.93NAGVAKK239 pKa = 10.75GNDD242 pKa = 3.26AALDD246 pKa = 3.71KK247 pKa = 11.1VNEE250 pKa = 4.44IIASIDD256 pKa = 3.34DD257 pKa = 3.97EE258 pKa = 4.5EE259 pKa = 5.03RR260 pKa = 11.84QDD262 pKa = 3.31MWSACMDD269 pKa = 3.82RR270 pKa = 11.84QPAA273 pKa = 3.25
MM1 pKa = 7.24MFLAGCGPAPQGSGDD16 pKa = 3.86AADD19 pKa = 4.81GSASASGILLGDD31 pKa = 4.09GKK33 pKa = 7.94TLRR36 pKa = 11.84VGMEE40 pKa = 3.77AAYY43 pKa = 10.13APYY46 pKa = 8.99NWQVSEE52 pKa = 4.13EE53 pKa = 4.44SEE55 pKa = 4.02FTIPIEE61 pKa = 4.2NVPGAYY67 pKa = 9.89ADD69 pKa = 4.36GYY71 pKa = 10.23DD72 pKa = 3.51VQVAKK77 pKa = 10.6IIADD81 pKa = 3.79GLGMEE86 pKa = 4.56PVAVKK91 pKa = 10.9LEE93 pKa = 4.07FGSLIDD99 pKa = 4.24ALNNGQVDD107 pKa = 3.91IVCAGMSVTPEE118 pKa = 3.58RR119 pKa = 11.84AAAAAFSDD127 pKa = 4.49SYY129 pKa = 11.82VDD131 pKa = 4.55DD132 pKa = 6.16DD133 pKa = 3.91IVMICRR139 pKa = 11.84ADD141 pKa = 3.26SAYY144 pKa = 10.71AGATTFDD151 pKa = 3.87EE152 pKa = 4.71LADD155 pKa = 3.96ASILGQAATMYY166 pKa = 10.76DD167 pKa = 3.67DD168 pKa = 5.47VIAQIEE174 pKa = 4.79GANHH178 pKa = 4.7MTPAEE183 pKa = 4.14TQPMVVEE190 pKa = 4.02NLRR193 pKa = 11.84NGTCDD198 pKa = 3.01IITYY202 pKa = 11.01SMLSAPKK209 pKa = 10.1LLEE212 pKa = 4.09VYY214 pKa = 9.98PDD216 pKa = 3.72LVVLDD221 pKa = 4.45MEE223 pKa = 4.95DD224 pKa = 3.42AFEE227 pKa = 4.72GSLMPDD233 pKa = 2.93NAGVAKK239 pKa = 10.75GNDD242 pKa = 3.26AALDD246 pKa = 3.71KK247 pKa = 11.1VNEE250 pKa = 4.44IIASIDD256 pKa = 3.34DD257 pKa = 3.97EE258 pKa = 4.5EE259 pKa = 5.03RR260 pKa = 11.84QDD262 pKa = 3.31MWSACMDD269 pKa = 3.82RR270 pKa = 11.84QPAA273 pKa = 3.25
Molecular weight: 28.7 kDa
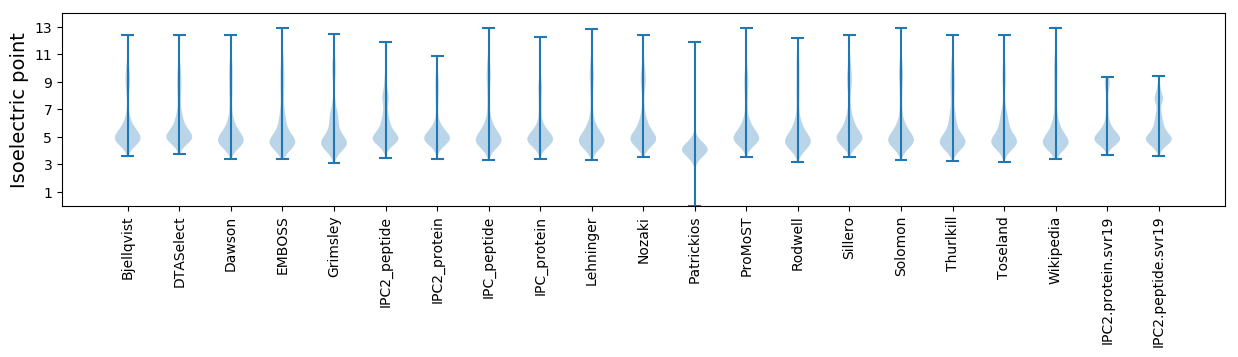
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A414NFS4|A0A414NFS4_9ACTN D-alanyl-lipoteichoic acid biosynthesis protein DltB OS=Collinsella intestinalis OX=147207 GN=dltB PE=3 SV=1
MM1 pKa = 7.38GALPGGGFMTEE12 pKa = 4.38EE13 pKa = 4.14KK14 pKa = 10.53QSTGLGDD21 pKa = 4.33GIARR25 pKa = 11.84VGDD28 pKa = 3.51IAQRR32 pKa = 11.84LCDD35 pKa = 3.35NRR37 pKa = 11.84RR38 pKa = 11.84FVIVSVCLLVFLVLLNRR55 pKa = 11.84VLAGEE60 pKa = 4.27IMALDD65 pKa = 4.16RR66 pKa = 11.84SAIALMVGHH75 pKa = 6.7VRR77 pKa = 11.84TAWLTPVMEE86 pKa = 4.56TVSFMVTPVPLFALIFAVALAGRR109 pKa = 11.84SRR111 pKa = 11.84GVGGLGWFCTINLAGSTILNQLLKK135 pKa = 10.52FAIQRR140 pKa = 11.84PRR142 pKa = 11.84PDD144 pKa = 2.76VSLRR148 pKa = 11.84LVEE151 pKa = 3.91IGGFSFPSGHH161 pKa = 6.22SMAAMAFFGLFIWLTWRR178 pKa = 11.84YY179 pKa = 9.56VRR181 pKa = 11.84DD182 pKa = 3.14RR183 pKa = 11.84RR184 pKa = 11.84ARR186 pKa = 11.84VGLTVFLCVMIAAVGFSRR204 pKa = 11.84VYY206 pKa = 10.87LGVHH210 pKa = 5.52YY211 pKa = 10.68ASDD214 pKa = 3.81VLGGFCASLIWLAVYY229 pKa = 8.62TKK231 pKa = 10.8VAGPTLAAAKK241 pKa = 10.11RR242 pKa = 11.84VGG244 pKa = 3.2
MM1 pKa = 7.38GALPGGGFMTEE12 pKa = 4.38EE13 pKa = 4.14KK14 pKa = 10.53QSTGLGDD21 pKa = 4.33GIARR25 pKa = 11.84VGDD28 pKa = 3.51IAQRR32 pKa = 11.84LCDD35 pKa = 3.35NRR37 pKa = 11.84RR38 pKa = 11.84FVIVSVCLLVFLVLLNRR55 pKa = 11.84VLAGEE60 pKa = 4.27IMALDD65 pKa = 4.16RR66 pKa = 11.84SAIALMVGHH75 pKa = 6.7VRR77 pKa = 11.84TAWLTPVMEE86 pKa = 4.56TVSFMVTPVPLFALIFAVALAGRR109 pKa = 11.84SRR111 pKa = 11.84GVGGLGWFCTINLAGSTILNQLLKK135 pKa = 10.52FAIQRR140 pKa = 11.84PRR142 pKa = 11.84PDD144 pKa = 2.76VSLRR148 pKa = 11.84LVEE151 pKa = 3.91IGGFSFPSGHH161 pKa = 6.22SMAAMAFFGLFIWLTWRR178 pKa = 11.84YY179 pKa = 9.56VRR181 pKa = 11.84DD182 pKa = 3.14RR183 pKa = 11.84RR184 pKa = 11.84ARR186 pKa = 11.84VGLTVFLCVMIAAVGFSRR204 pKa = 11.84VYY206 pKa = 10.87LGVHH210 pKa = 5.52YY211 pKa = 10.68ASDD214 pKa = 3.81VLGGFCASLIWLAVYY229 pKa = 8.62TKK231 pKa = 10.8VAGPTLAAAKK241 pKa = 10.11RR242 pKa = 11.84VGG244 pKa = 3.2
Molecular weight: 26.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
497327 |
24 |
3859 |
347.8 |
37.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.637 ± 0.086 | 1.521 ± 0.03 |
6.282 ± 0.05 | 6.845 ± 0.07 |
3.667 ± 0.04 | 8.291 ± 0.055 |
1.969 ± 0.027 | 5.325 ± 0.056 |
3.549 ± 0.054 | 9.26 ± 0.072 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.841 ± 0.031 | 2.781 ± 0.043 |
4.364 ± 0.039 | 2.686 ± 0.028 |
6.338 ± 0.073 | 5.707 ± 0.043 |
5.234 ± 0.066 | 8.092 ± 0.056 |
0.959 ± 0.022 | 2.651 ± 0.036 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
