
Chitinophaga rupis
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Chitinophagia; Chitinophagales; Chitinophagaceae; Chitinophaga
Average proteome isoelectric point is 6.88
Get precalculated fractions of proteins
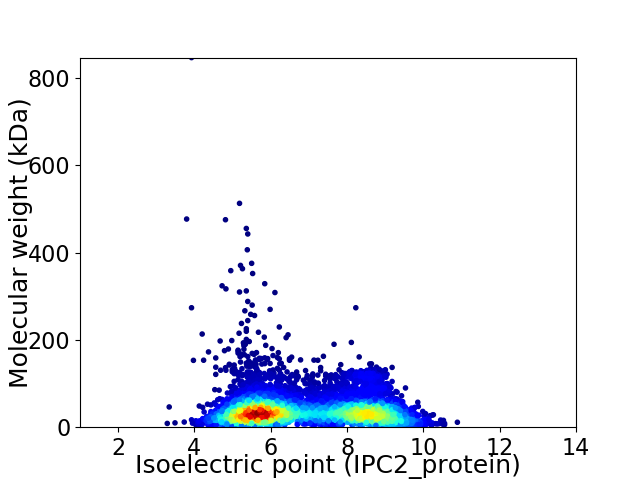
Virtual 2D-PAGE plot for 6498 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
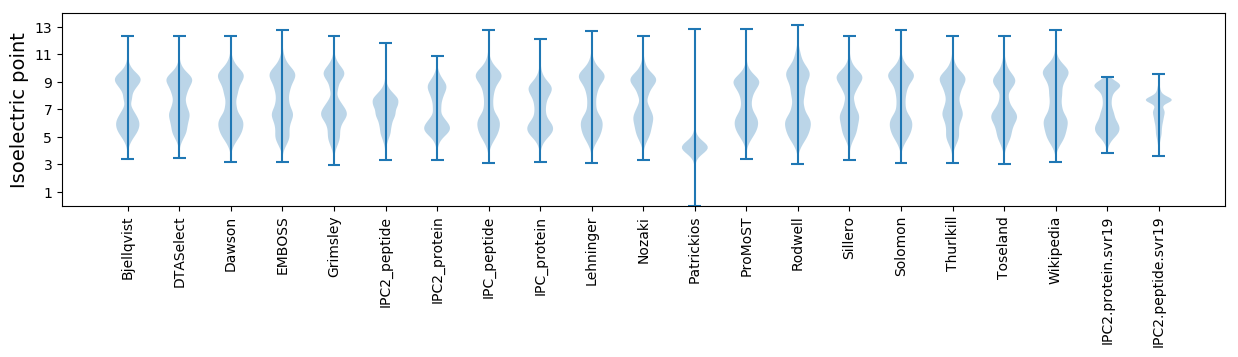
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H7RNU7|A0A1H7RNU7_9BACT Peptidase family S41 OS=Chitinophaga rupis OX=573321 GN=SAMN04488505_102673 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 10.41ALYY5 pKa = 10.26LLLAGLLFGFAATAQYY21 pKa = 10.81TSTTINSGVSQCALAKK37 pKa = 10.51DD38 pKa = 3.48IAGNIYY44 pKa = 10.52AVRR47 pKa = 11.84TDD49 pKa = 3.89PNDD52 pKa = 3.26ATKK55 pKa = 9.27YY56 pKa = 8.82TVVKK60 pKa = 9.53YY61 pKa = 10.86VNGAGAPVRR70 pKa = 11.84LNASLQLVTSGIEE83 pKa = 4.23YY84 pKa = 9.22PWGLAVNSLGDD95 pKa = 3.56VYY97 pKa = 10.56VTNPEE102 pKa = 4.21GADD105 pKa = 2.83GWEE108 pKa = 4.4IIKK111 pKa = 8.9LTAPTYY117 pKa = 7.87TATVIQSGKK126 pKa = 9.81FYY128 pKa = 11.04SSLAIDD134 pKa = 3.73QNNDD138 pKa = 2.98IITTEE143 pKa = 4.22YY144 pKa = 10.18IDD146 pKa = 4.94PDD148 pKa = 3.64GTFNNGDD155 pKa = 3.29DD156 pKa = 3.62KK157 pKa = 11.63YY158 pKa = 11.14RR159 pKa = 11.84VVKK162 pKa = 10.33YY163 pKa = 10.36LYY165 pKa = 10.01TNEE168 pKa = 4.09TGAGTVLYY176 pKa = 10.79AGVPYY181 pKa = 10.37EE182 pKa = 4.7NDD184 pKa = 2.99FSYY187 pKa = 10.12PWSIVVDD194 pKa = 4.07GFGNIYY200 pKa = 10.56FLDD203 pKa = 4.23FVTNDD208 pKa = 4.15PAPGTANDD216 pKa = 3.84GGQLIKK222 pKa = 10.08LTYY225 pKa = 8.19PAYY228 pKa = 9.42SASVINSGKK237 pKa = 10.6GISTLALDD245 pKa = 4.9ASNNLYY251 pKa = 9.15TVEE254 pKa = 4.58AVDD257 pKa = 3.85PTTSHH262 pKa = 4.91VVKK265 pKa = 10.61YY266 pKa = 9.76SYY268 pKa = 10.57PVTAGASGTAITPVTLVSAAGFYY291 pKa = 9.69PWGMVIKK298 pKa = 10.49GSSIFVNDD306 pKa = 3.48ATDD309 pKa = 4.01DD310 pKa = 4.02PSTPGDD316 pKa = 3.6PADD319 pKa = 3.88GAFKK323 pKa = 10.77RR324 pKa = 11.84LDD326 pKa = 3.72PPTIMVNSVNRR337 pKa = 11.84VTATPTKK344 pKa = 9.75LASVQFTVTFSGSATGVTTSAFTLTTTGLTGASITSVSGSGTTYY388 pKa = 10.12TVTVNTGTGDD398 pKa = 3.2GTLRR402 pKa = 11.84LDD404 pKa = 3.65VNGTGISPTVSPAPFITGQVYY425 pKa = 9.03TIDD428 pKa = 3.38KK429 pKa = 7.52TAPTTTITATPPVLTNSSSGTFTFTSNEE457 pKa = 3.77AGTFEE462 pKa = 4.53ASLDD466 pKa = 3.79GGPYY470 pKa = 8.76TAATSPRR477 pKa = 11.84TYY479 pKa = 10.3TGLADD484 pKa = 4.1GSHH487 pKa = 5.9TFSVRR492 pKa = 11.84AIDD495 pKa = 3.46AAGNVAVTPASYY507 pKa = 8.82TWTVDD512 pKa = 2.93QTAPNTTITSAPPAFTNSSNATFTFISSEE541 pKa = 4.05AGSTFQASLDD551 pKa = 3.87GGPFTPVTSPSTYY564 pKa = 9.85TGLADD569 pKa = 4.04GSHH572 pKa = 5.9TFSVRR577 pKa = 11.84AIDD580 pKa = 3.75PAGNTDD586 pKa = 3.94ATPDD590 pKa = 3.69SYY592 pKa = 11.48PWTIDD597 pKa = 2.91QTAPTVTSVGVPANDD612 pKa = 3.99YY613 pKa = 10.34YY614 pKa = 11.3HH615 pKa = 7.42AGQNLDD621 pKa = 3.35FTVNFSEE628 pKa = 4.1NVTVTGTPYY637 pKa = 10.57INLTVGSTGLQAAYY651 pKa = 10.72VSGSGTNALVFRR663 pKa = 11.84YY664 pKa = 7.29TVQSGDD670 pKa = 3.21ADD672 pKa = 3.91ADD674 pKa = 3.95GVTLGTLALNGGTIQDD690 pKa = 3.95AATNDD695 pKa = 3.5ANVTLNGVPSLANVRR710 pKa = 11.84VNTTIPSVVLSTTASSPRR728 pKa = 11.84NSGFTITLTFSEE740 pKa = 4.91AVTGLSVADD749 pKa = 4.03LNVLNTTVSNLQTADD764 pKa = 3.58NITYY768 pKa = 10.36TMDD771 pKa = 3.9LQPSVNGVINMQLPANAVQNIGHH794 pKa = 6.38NPNTASNTLSITYY807 pKa = 9.72DD808 pKa = 3.81AIAPVVTSVSVPANGYY824 pKa = 9.49YY825 pKa = 10.07KK826 pKa = 10.81ASDD829 pKa = 3.29ALTFTVNFSEE839 pKa = 4.66NITVAGIPQLSMIIGATPRR858 pKa = 11.84QAAYY862 pKa = 10.87VSGNGTNALTFTYY875 pKa = 9.2TVQSGEE881 pKa = 3.99LDD883 pKa = 3.21ADD885 pKa = 4.89GIAVTSLSANGGSLQDD901 pKa = 3.63AATNNANLTLNSVGSTTGVLVDD923 pKa = 4.41AVAPTVTSVAVPANDD938 pKa = 3.87YY939 pKa = 10.06YY940 pKa = 11.7HH941 pKa = 7.0NGQNLDD947 pKa = 3.43FTVNFSEE954 pKa = 4.1NVTVTGTPYY963 pKa = 10.57INLTVGSTGRR973 pKa = 11.84QAAYY977 pKa = 10.69VSGSGTNALVFRR989 pKa = 11.84YY990 pKa = 7.36TVQLSDD996 pKa = 4.21ADD998 pKa = 3.57NDD1000 pKa = 4.83GIQLGTLALNGGPIQDD1016 pKa = 3.45VAGNNAVLTLNGVPSLANVRR1036 pKa = 11.84VNNIIPTVSITSAAPDD1052 pKa = 3.87PLNTGFTATITFSEE1066 pKa = 4.57AVTGLVAGDD1075 pKa = 3.68LTSINATASNLQTSDD1090 pKa = 3.51NITYY1094 pKa = 9.14TVDD1097 pKa = 3.78LAPAADD1103 pKa = 3.98GTVSLQVPGNAAEE1116 pKa = 4.51NIGHH1120 pKa = 6.71NGNTASNTLTRR1131 pKa = 11.84TYY1133 pKa = 10.91DD1134 pKa = 3.05ATAPTVTSVSVPANGYY1150 pKa = 9.27YY1151 pKa = 10.4KK1152 pKa = 10.79AGDD1155 pKa = 3.49ALTFTVNFSEE1165 pKa = 4.66NITVAGIPQLSMIIGAMPRR1184 pKa = 11.84QAAYY1188 pKa = 10.9VSGNGTNALTFTYY1201 pKa = 9.2TVQSGEE1207 pKa = 3.99LDD1209 pKa = 3.21ADD1211 pKa = 4.89GIAVTSLSANGGSLMDD1227 pKa = 4.4AATNNANLTLNSVGSTTGVLVDD1249 pKa = 4.3AVAPAVTSVGVPANGYY1265 pKa = 8.19YY1266 pKa = 8.05QTGQNLDD1273 pKa = 3.46FTVNFSEE1280 pKa = 4.1NVTVTGTPTVPITIGSATVDD1300 pKa = 3.02ASYY1303 pKa = 11.01ISGSGTSALVFRR1315 pKa = 11.84YY1316 pKa = 7.21TVQAGEE1322 pKa = 3.96QDD1324 pKa = 3.36LDD1326 pKa = 4.51GISVGSTIALNGGTIRR1342 pKa = 11.84DD1343 pKa = 3.64AATNNAVLTLNSIGSTANVFVYY1365 pKa = 10.39SVVPGVTISTTAPALVNAGFTLTVTFTEE1393 pKa = 4.39AVTGFTASDD1402 pKa = 3.13ITATNATVSNLATSDD1417 pKa = 3.88NITYY1421 pKa = 9.3TLDD1424 pKa = 3.17VTPAADD1430 pKa = 3.63GAVSLQVPASSAVNIAGNGNTASNTLSLTYY1460 pKa = 10.4DD1461 pKa = 3.03ATAPVVTTVAVPPNGYY1477 pKa = 9.78YY1478 pKa = 8.7KK1479 pKa = 10.54TGGILNFTINFSEE1492 pKa = 4.16NVTVTGTPQLSMVIGSTIRR1511 pKa = 11.84YY1512 pKa = 8.42ANYY1515 pKa = 9.56ISGSGTGALVFRR1527 pKa = 11.84YY1528 pKa = 7.07TVQNGEE1534 pKa = 3.77EE1535 pKa = 4.2DD1536 pKa = 3.53MNGITLGTLQLNGGTIKK1553 pKa = 10.87DD1554 pKa = 3.57AATNDD1559 pKa = 3.47AVLTLNGVPSTTNVRR1574 pKa = 11.84VHH1576 pKa = 5.73TAVPSVTLTTAAPALVNAPFTITATFSEE1604 pKa = 4.74SMAGVALADD1613 pKa = 3.57FTVTNGTVSSLSGTANTVFTLTVTPTADD1641 pKa = 3.52GPVSITFPANKK1652 pKa = 9.36ALNVAGTNNTASNTLSFTYY1671 pKa = 10.37DD1672 pKa = 2.75GTAPVITAVSVPPNGYY1688 pKa = 10.02YY1689 pKa = 10.47NATGVLSFTATFSEE1703 pKa = 4.39NVTVTGTPQLGVIIGATTQQADD1725 pKa = 4.04YY1726 pKa = 11.2VSGSGTNQLTFSYY1739 pKa = 9.86TVQTGDD1745 pKa = 3.28NDD1747 pKa = 3.35MDD1749 pKa = 5.29GIALGNLTLNGGTIQDD1765 pKa = 3.64AATNNAVLTLNGVPSTAGIFVNTTAPTVVVSTTAVSPGNQPFTATFTFSEE1815 pKa = 4.77KK1816 pKa = 9.21VTGFTSSDD1824 pKa = 2.8IVVTNGAAGATSTSDD1839 pKa = 3.28NITYY1843 pKa = 7.79TALITPAADD1852 pKa = 3.28GTVTVQVPANVAVNIGNNGNVASNTITYY1880 pKa = 8.54TYY1882 pKa = 10.64DD1883 pKa = 2.83GTAPAITSVAVPPNGTYY1900 pKa = 10.23NASDD1904 pKa = 3.5VLNFIVNYY1912 pKa = 9.89DD1913 pKa = 3.58EE1914 pKa = 4.16NVNVIGAGGIPSLNLTIGSSAVQAAYY1940 pKa = 9.82ISGSGTNALTFSYY1953 pKa = 9.49TVQAGEE1959 pKa = 4.0QDD1961 pKa = 3.47LDD1963 pKa = 4.58GITIAPTMIGTMKK1976 pKa = 10.71DD1977 pKa = 3.3DD1978 pKa = 4.6AGNTASNTLNGVGNTSGVLVYY1999 pKa = 9.44TATPTVQLSGTVQANAPFTLTITFSEE2025 pKa = 4.48AVTGFTLSDD2034 pKa = 3.11ITATNATLTNLNTTDD2049 pKa = 3.86NITYY2053 pKa = 7.37TALVTPVADD2062 pKa = 3.82GLVTLQVPANAAVNIVSNGNTASNTISYY2090 pKa = 8.76TYY2092 pKa = 10.71DD2093 pKa = 2.64ATAPVITSVDD2103 pKa = 3.61VPANAYY2109 pKa = 7.58YY2110 pKa = 7.75TTGQNLDD2117 pKa = 3.76FTVHH2121 pKa = 5.42MSEE2124 pKa = 4.2AVNVAGGTPSIPVTIGATTVQANYY2148 pKa = 10.6VSGTGTNALLFRR2160 pKa = 11.84YY2161 pKa = 7.24TVLNGQMDD2169 pKa = 3.93MDD2171 pKa = 5.55GISLGAALALNGSTLRR2187 pKa = 11.84DD2188 pKa = 3.37PAGNNAVLTLNSVGSTTGVRR2208 pKa = 11.84VNTAHH2213 pKa = 6.45PTVVVATTVATRR2225 pKa = 11.84VNTAFNVTLTFSEE2238 pKa = 4.58AVTGLTTTGITATNATVSNLNTTDD2262 pKa = 3.71NITYY2266 pKa = 7.55TATITPVADD2275 pKa = 3.42GAVNVSVPANAAVNTGNNGNTASNTVTITYY2305 pKa = 10.19DD2306 pKa = 2.98VTAPVIANASFNVYY2320 pKa = 10.6DD2321 pKa = 3.93NSTAGTVVGTLTAAEE2336 pKa = 4.23TAGTLQSWTLVTDD2349 pKa = 4.46GSGGAFALNAATGAITVKK2367 pKa = 10.6DD2368 pKa = 3.41AALLKK2373 pKa = 9.58TKK2375 pKa = 10.45AGQTFTLTVTVSDD2388 pKa = 4.37GLNTSAATPVTVKK2401 pKa = 10.78VLIAFINKK2409 pKa = 9.4APTLDD2414 pKa = 4.62AIADD2418 pKa = 3.77AKK2420 pKa = 11.06VCTGTDD2426 pKa = 2.81THH2428 pKa = 7.02TIQLTGASATEE2439 pKa = 3.93AGQTYY2444 pKa = 9.01TITAVSDD2451 pKa = 3.52QPFFDD2456 pKa = 4.02ALSVNASNILSYY2468 pKa = 10.81KK2469 pKa = 10.17LKK2471 pKa = 10.89ASVTTGTATITVTIKK2486 pKa = 11.16DD2487 pKa = 3.43NGGTDD2492 pKa = 3.11NGGVDD2497 pKa = 3.32TLRR2500 pKa = 11.84RR2501 pKa = 11.84SFTITVNEE2509 pKa = 4.48LPSISISSDD2518 pKa = 2.72KK2519 pKa = 11.07GSTISKK2525 pKa = 10.27GDD2527 pKa = 3.8VIHH2530 pKa = 7.31LNATRR2535 pKa = 11.84VNGYY2539 pKa = 7.65TYY2541 pKa = 10.59SWSPADD2547 pKa = 5.32GILSGQNNFILEE2559 pKa = 4.18ARR2561 pKa = 11.84PLSNITYY2568 pKa = 9.87VVTATTAAGCSSTANIAITVVEE2590 pKa = 4.58DD2591 pKa = 4.14FKK2593 pKa = 11.83VDD2595 pKa = 4.54AINILTPNGDD2605 pKa = 3.12GRR2607 pKa = 11.84NDD2609 pKa = 2.64KK2610 pKa = 10.08WVIRR2614 pKa = 11.84NLDD2617 pKa = 3.79SYY2619 pKa = 10.91PDD2621 pKa = 3.67NEE2623 pKa = 3.94VSIFDD2628 pKa = 3.38RR2629 pKa = 11.84TGRR2632 pKa = 11.84MIYY2635 pKa = 10.23HH2636 pKa = 6.84RR2637 pKa = 11.84VNYY2640 pKa = 10.16SNDD2643 pKa = 2.88WDD2645 pKa = 3.76ATLNGSPLAEE2655 pKa = 3.64GTYY2658 pKa = 10.7YY2659 pKa = 10.89YY2660 pKa = 10.82VLKK2663 pKa = 10.73INGGAKK2669 pKa = 7.79TAKK2672 pKa = 10.24GYY2674 pKa = 8.61ITIIRR2679 pKa = 11.84DD2680 pKa = 3.18QHH2682 pKa = 5.62
MM1 pKa = 7.35KK2 pKa = 10.41ALYY5 pKa = 10.26LLLAGLLFGFAATAQYY21 pKa = 10.81TSTTINSGVSQCALAKK37 pKa = 10.51DD38 pKa = 3.48IAGNIYY44 pKa = 10.52AVRR47 pKa = 11.84TDD49 pKa = 3.89PNDD52 pKa = 3.26ATKK55 pKa = 9.27YY56 pKa = 8.82TVVKK60 pKa = 9.53YY61 pKa = 10.86VNGAGAPVRR70 pKa = 11.84LNASLQLVTSGIEE83 pKa = 4.23YY84 pKa = 9.22PWGLAVNSLGDD95 pKa = 3.56VYY97 pKa = 10.56VTNPEE102 pKa = 4.21GADD105 pKa = 2.83GWEE108 pKa = 4.4IIKK111 pKa = 8.9LTAPTYY117 pKa = 7.87TATVIQSGKK126 pKa = 9.81FYY128 pKa = 11.04SSLAIDD134 pKa = 3.73QNNDD138 pKa = 2.98IITTEE143 pKa = 4.22YY144 pKa = 10.18IDD146 pKa = 4.94PDD148 pKa = 3.64GTFNNGDD155 pKa = 3.29DD156 pKa = 3.62KK157 pKa = 11.63YY158 pKa = 11.14RR159 pKa = 11.84VVKK162 pKa = 10.33YY163 pKa = 10.36LYY165 pKa = 10.01TNEE168 pKa = 4.09TGAGTVLYY176 pKa = 10.79AGVPYY181 pKa = 10.37EE182 pKa = 4.7NDD184 pKa = 2.99FSYY187 pKa = 10.12PWSIVVDD194 pKa = 4.07GFGNIYY200 pKa = 10.56FLDD203 pKa = 4.23FVTNDD208 pKa = 4.15PAPGTANDD216 pKa = 3.84GGQLIKK222 pKa = 10.08LTYY225 pKa = 8.19PAYY228 pKa = 9.42SASVINSGKK237 pKa = 10.6GISTLALDD245 pKa = 4.9ASNNLYY251 pKa = 9.15TVEE254 pKa = 4.58AVDD257 pKa = 3.85PTTSHH262 pKa = 4.91VVKK265 pKa = 10.61YY266 pKa = 9.76SYY268 pKa = 10.57PVTAGASGTAITPVTLVSAAGFYY291 pKa = 9.69PWGMVIKK298 pKa = 10.49GSSIFVNDD306 pKa = 3.48ATDD309 pKa = 4.01DD310 pKa = 4.02PSTPGDD316 pKa = 3.6PADD319 pKa = 3.88GAFKK323 pKa = 10.77RR324 pKa = 11.84LDD326 pKa = 3.72PPTIMVNSVNRR337 pKa = 11.84VTATPTKK344 pKa = 9.75LASVQFTVTFSGSATGVTTSAFTLTTTGLTGASITSVSGSGTTYY388 pKa = 10.12TVTVNTGTGDD398 pKa = 3.2GTLRR402 pKa = 11.84LDD404 pKa = 3.65VNGTGISPTVSPAPFITGQVYY425 pKa = 9.03TIDD428 pKa = 3.38KK429 pKa = 7.52TAPTTTITATPPVLTNSSSGTFTFTSNEE457 pKa = 3.77AGTFEE462 pKa = 4.53ASLDD466 pKa = 3.79GGPYY470 pKa = 8.76TAATSPRR477 pKa = 11.84TYY479 pKa = 10.3TGLADD484 pKa = 4.1GSHH487 pKa = 5.9TFSVRR492 pKa = 11.84AIDD495 pKa = 3.46AAGNVAVTPASYY507 pKa = 8.82TWTVDD512 pKa = 2.93QTAPNTTITSAPPAFTNSSNATFTFISSEE541 pKa = 4.05AGSTFQASLDD551 pKa = 3.87GGPFTPVTSPSTYY564 pKa = 9.85TGLADD569 pKa = 4.04GSHH572 pKa = 5.9TFSVRR577 pKa = 11.84AIDD580 pKa = 3.75PAGNTDD586 pKa = 3.94ATPDD590 pKa = 3.69SYY592 pKa = 11.48PWTIDD597 pKa = 2.91QTAPTVTSVGVPANDD612 pKa = 3.99YY613 pKa = 10.34YY614 pKa = 11.3HH615 pKa = 7.42AGQNLDD621 pKa = 3.35FTVNFSEE628 pKa = 4.1NVTVTGTPYY637 pKa = 10.57INLTVGSTGLQAAYY651 pKa = 10.72VSGSGTNALVFRR663 pKa = 11.84YY664 pKa = 7.29TVQSGDD670 pKa = 3.21ADD672 pKa = 3.91ADD674 pKa = 3.95GVTLGTLALNGGTIQDD690 pKa = 3.95AATNDD695 pKa = 3.5ANVTLNGVPSLANVRR710 pKa = 11.84VNTTIPSVVLSTTASSPRR728 pKa = 11.84NSGFTITLTFSEE740 pKa = 4.91AVTGLSVADD749 pKa = 4.03LNVLNTTVSNLQTADD764 pKa = 3.58NITYY768 pKa = 10.36TMDD771 pKa = 3.9LQPSVNGVINMQLPANAVQNIGHH794 pKa = 6.38NPNTASNTLSITYY807 pKa = 9.72DD808 pKa = 3.81AIAPVVTSVSVPANGYY824 pKa = 9.49YY825 pKa = 10.07KK826 pKa = 10.81ASDD829 pKa = 3.29ALTFTVNFSEE839 pKa = 4.66NITVAGIPQLSMIIGATPRR858 pKa = 11.84QAAYY862 pKa = 10.87VSGNGTNALTFTYY875 pKa = 9.2TVQSGEE881 pKa = 3.99LDD883 pKa = 3.21ADD885 pKa = 4.89GIAVTSLSANGGSLQDD901 pKa = 3.63AATNNANLTLNSVGSTTGVLVDD923 pKa = 4.41AVAPTVTSVAVPANDD938 pKa = 3.87YY939 pKa = 10.06YY940 pKa = 11.7HH941 pKa = 7.0NGQNLDD947 pKa = 3.43FTVNFSEE954 pKa = 4.1NVTVTGTPYY963 pKa = 10.57INLTVGSTGRR973 pKa = 11.84QAAYY977 pKa = 10.69VSGSGTNALVFRR989 pKa = 11.84YY990 pKa = 7.36TVQLSDD996 pKa = 4.21ADD998 pKa = 3.57NDD1000 pKa = 4.83GIQLGTLALNGGPIQDD1016 pKa = 3.45VAGNNAVLTLNGVPSLANVRR1036 pKa = 11.84VNNIIPTVSITSAAPDD1052 pKa = 3.87PLNTGFTATITFSEE1066 pKa = 4.57AVTGLVAGDD1075 pKa = 3.68LTSINATASNLQTSDD1090 pKa = 3.51NITYY1094 pKa = 9.14TVDD1097 pKa = 3.78LAPAADD1103 pKa = 3.98GTVSLQVPGNAAEE1116 pKa = 4.51NIGHH1120 pKa = 6.71NGNTASNTLTRR1131 pKa = 11.84TYY1133 pKa = 10.91DD1134 pKa = 3.05ATAPTVTSVSVPANGYY1150 pKa = 9.27YY1151 pKa = 10.4KK1152 pKa = 10.79AGDD1155 pKa = 3.49ALTFTVNFSEE1165 pKa = 4.66NITVAGIPQLSMIIGAMPRR1184 pKa = 11.84QAAYY1188 pKa = 10.9VSGNGTNALTFTYY1201 pKa = 9.2TVQSGEE1207 pKa = 3.99LDD1209 pKa = 3.21ADD1211 pKa = 4.89GIAVTSLSANGGSLMDD1227 pKa = 4.4AATNNANLTLNSVGSTTGVLVDD1249 pKa = 4.3AVAPAVTSVGVPANGYY1265 pKa = 8.19YY1266 pKa = 8.05QTGQNLDD1273 pKa = 3.46FTVNFSEE1280 pKa = 4.1NVTVTGTPTVPITIGSATVDD1300 pKa = 3.02ASYY1303 pKa = 11.01ISGSGTSALVFRR1315 pKa = 11.84YY1316 pKa = 7.21TVQAGEE1322 pKa = 3.96QDD1324 pKa = 3.36LDD1326 pKa = 4.51GISVGSTIALNGGTIRR1342 pKa = 11.84DD1343 pKa = 3.64AATNNAVLTLNSIGSTANVFVYY1365 pKa = 10.39SVVPGVTISTTAPALVNAGFTLTVTFTEE1393 pKa = 4.39AVTGFTASDD1402 pKa = 3.13ITATNATVSNLATSDD1417 pKa = 3.88NITYY1421 pKa = 9.3TLDD1424 pKa = 3.17VTPAADD1430 pKa = 3.63GAVSLQVPASSAVNIAGNGNTASNTLSLTYY1460 pKa = 10.4DD1461 pKa = 3.03ATAPVVTTVAVPPNGYY1477 pKa = 9.78YY1478 pKa = 8.7KK1479 pKa = 10.54TGGILNFTINFSEE1492 pKa = 4.16NVTVTGTPQLSMVIGSTIRR1511 pKa = 11.84YY1512 pKa = 8.42ANYY1515 pKa = 9.56ISGSGTGALVFRR1527 pKa = 11.84YY1528 pKa = 7.07TVQNGEE1534 pKa = 3.77EE1535 pKa = 4.2DD1536 pKa = 3.53MNGITLGTLQLNGGTIKK1553 pKa = 10.87DD1554 pKa = 3.57AATNDD1559 pKa = 3.47AVLTLNGVPSTTNVRR1574 pKa = 11.84VHH1576 pKa = 5.73TAVPSVTLTTAAPALVNAPFTITATFSEE1604 pKa = 4.74SMAGVALADD1613 pKa = 3.57FTVTNGTVSSLSGTANTVFTLTVTPTADD1641 pKa = 3.52GPVSITFPANKK1652 pKa = 9.36ALNVAGTNNTASNTLSFTYY1671 pKa = 10.37DD1672 pKa = 2.75GTAPVITAVSVPPNGYY1688 pKa = 10.02YY1689 pKa = 10.47NATGVLSFTATFSEE1703 pKa = 4.39NVTVTGTPQLGVIIGATTQQADD1725 pKa = 4.04YY1726 pKa = 11.2VSGSGTNQLTFSYY1739 pKa = 9.86TVQTGDD1745 pKa = 3.28NDD1747 pKa = 3.35MDD1749 pKa = 5.29GIALGNLTLNGGTIQDD1765 pKa = 3.64AATNNAVLTLNGVPSTAGIFVNTTAPTVVVSTTAVSPGNQPFTATFTFSEE1815 pKa = 4.77KK1816 pKa = 9.21VTGFTSSDD1824 pKa = 2.8IVVTNGAAGATSTSDD1839 pKa = 3.28NITYY1843 pKa = 7.79TALITPAADD1852 pKa = 3.28GTVTVQVPANVAVNIGNNGNVASNTITYY1880 pKa = 8.54TYY1882 pKa = 10.64DD1883 pKa = 2.83GTAPAITSVAVPPNGTYY1900 pKa = 10.23NASDD1904 pKa = 3.5VLNFIVNYY1912 pKa = 9.89DD1913 pKa = 3.58EE1914 pKa = 4.16NVNVIGAGGIPSLNLTIGSSAVQAAYY1940 pKa = 9.82ISGSGTNALTFSYY1953 pKa = 9.49TVQAGEE1959 pKa = 4.0QDD1961 pKa = 3.47LDD1963 pKa = 4.58GITIAPTMIGTMKK1976 pKa = 10.71DD1977 pKa = 3.3DD1978 pKa = 4.6AGNTASNTLNGVGNTSGVLVYY1999 pKa = 9.44TATPTVQLSGTVQANAPFTLTITFSEE2025 pKa = 4.48AVTGFTLSDD2034 pKa = 3.11ITATNATLTNLNTTDD2049 pKa = 3.86NITYY2053 pKa = 7.37TALVTPVADD2062 pKa = 3.82GLVTLQVPANAAVNIVSNGNTASNTISYY2090 pKa = 8.76TYY2092 pKa = 10.71DD2093 pKa = 2.64ATAPVITSVDD2103 pKa = 3.61VPANAYY2109 pKa = 7.58YY2110 pKa = 7.75TTGQNLDD2117 pKa = 3.76FTVHH2121 pKa = 5.42MSEE2124 pKa = 4.2AVNVAGGTPSIPVTIGATTVQANYY2148 pKa = 10.6VSGTGTNALLFRR2160 pKa = 11.84YY2161 pKa = 7.24TVLNGQMDD2169 pKa = 3.93MDD2171 pKa = 5.55GISLGAALALNGSTLRR2187 pKa = 11.84DD2188 pKa = 3.37PAGNNAVLTLNSVGSTTGVRR2208 pKa = 11.84VNTAHH2213 pKa = 6.45PTVVVATTVATRR2225 pKa = 11.84VNTAFNVTLTFSEE2238 pKa = 4.58AVTGLTTTGITATNATVSNLNTTDD2262 pKa = 3.71NITYY2266 pKa = 7.55TATITPVADD2275 pKa = 3.42GAVNVSVPANAAVNTGNNGNTASNTVTITYY2305 pKa = 10.19DD2306 pKa = 2.98VTAPVIANASFNVYY2320 pKa = 10.6DD2321 pKa = 3.93NSTAGTVVGTLTAAEE2336 pKa = 4.23TAGTLQSWTLVTDD2349 pKa = 4.46GSGGAFALNAATGAITVKK2367 pKa = 10.6DD2368 pKa = 3.41AALLKK2373 pKa = 9.58TKK2375 pKa = 10.45AGQTFTLTVTVSDD2388 pKa = 4.37GLNTSAATPVTVKK2401 pKa = 10.78VLIAFINKK2409 pKa = 9.4APTLDD2414 pKa = 4.62AIADD2418 pKa = 3.77AKK2420 pKa = 11.06VCTGTDD2426 pKa = 2.81THH2428 pKa = 7.02TIQLTGASATEE2439 pKa = 3.93AGQTYY2444 pKa = 9.01TITAVSDD2451 pKa = 3.52QPFFDD2456 pKa = 4.02ALSVNASNILSYY2468 pKa = 10.81KK2469 pKa = 10.17LKK2471 pKa = 10.89ASVTTGTATITVTIKK2486 pKa = 11.16DD2487 pKa = 3.43NGGTDD2492 pKa = 3.11NGGVDD2497 pKa = 3.32TLRR2500 pKa = 11.84RR2501 pKa = 11.84SFTITVNEE2509 pKa = 4.48LPSISISSDD2518 pKa = 2.72KK2519 pKa = 11.07GSTISKK2525 pKa = 10.27GDD2527 pKa = 3.8VIHH2530 pKa = 7.31LNATRR2535 pKa = 11.84VNGYY2539 pKa = 7.65TYY2541 pKa = 10.59SWSPADD2547 pKa = 5.32GILSGQNNFILEE2559 pKa = 4.18ARR2561 pKa = 11.84PLSNITYY2568 pKa = 9.87VVTATTAAGCSSTANIAITVVEE2590 pKa = 4.58DD2591 pKa = 4.14FKK2593 pKa = 11.83VDD2595 pKa = 4.54AINILTPNGDD2605 pKa = 3.12GRR2607 pKa = 11.84NDD2609 pKa = 2.64KK2610 pKa = 10.08WVIRR2614 pKa = 11.84NLDD2617 pKa = 3.79SYY2619 pKa = 10.91PDD2621 pKa = 3.67NEE2623 pKa = 3.94VSIFDD2628 pKa = 3.38RR2629 pKa = 11.84TGRR2632 pKa = 11.84MIYY2635 pKa = 10.23HH2636 pKa = 6.84RR2637 pKa = 11.84VNYY2640 pKa = 10.16SNDD2643 pKa = 2.88WDD2645 pKa = 3.76ATLNGSPLAEE2655 pKa = 3.64GTYY2658 pKa = 10.7YY2659 pKa = 10.89YY2660 pKa = 10.82VLKK2663 pKa = 10.73INGGAKK2669 pKa = 7.79TAKK2672 pKa = 10.24GYY2674 pKa = 8.61ITIIRR2679 pKa = 11.84DD2680 pKa = 3.18QHH2682 pKa = 5.62
Molecular weight: 273.69 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H7T0Y0|A0A1H7T0Y0_9BACT P4Hc domain-containing protein OS=Chitinophaga rupis OX=573321 GN=SAMN04488505_1021100 PE=4 SV=1
MM1 pKa = 6.94MAHH4 pKa = 5.81VQLSDD9 pKa = 3.14KK10 pKa = 10.48KK11 pKa = 9.51VRR13 pKa = 11.84SLIRR17 pKa = 11.84SGNIRR22 pKa = 11.84FGGNKK27 pKa = 9.44KK28 pKa = 10.19LRR30 pKa = 11.84IYY32 pKa = 11.02GLLSCVSGKK41 pKa = 7.54QMKK44 pKa = 9.42AANRR48 pKa = 11.84TFFATEE54 pKa = 3.65QEE56 pKa = 4.5AIAAGYY62 pKa = 8.66RR63 pKa = 11.84PCGHH67 pKa = 6.94CMPALYY73 pKa = 10.04RR74 pKa = 11.84AWKK77 pKa = 10.0AGAIPAHH84 pKa = 5.93GRR86 pKa = 3.35
MM1 pKa = 6.94MAHH4 pKa = 5.81VQLSDD9 pKa = 3.14KK10 pKa = 10.48KK11 pKa = 9.51VRR13 pKa = 11.84SLIRR17 pKa = 11.84SGNIRR22 pKa = 11.84FGGNKK27 pKa = 9.44KK28 pKa = 10.19LRR30 pKa = 11.84IYY32 pKa = 11.02GLLSCVSGKK41 pKa = 7.54QMKK44 pKa = 9.42AANRR48 pKa = 11.84TFFATEE54 pKa = 3.65QEE56 pKa = 4.5AIAAGYY62 pKa = 8.66RR63 pKa = 11.84PCGHH67 pKa = 6.94CMPALYY73 pKa = 10.04RR74 pKa = 11.84AWKK77 pKa = 10.0AGAIPAHH84 pKa = 5.93GRR86 pKa = 3.35
Molecular weight: 9.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2498919 |
29 |
8261 |
384.6 |
42.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.466 ± 0.034 | 0.862 ± 0.011 |
5.128 ± 0.022 | 5.134 ± 0.029 |
4.52 ± 0.024 | 7.122 ± 0.031 |
2.072 ± 0.016 | 6.419 ± 0.024 |
5.815 ± 0.031 | 9.653 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.347 ± 0.016 | 5.181 ± 0.03 |
4.348 ± 0.019 | 4.417 ± 0.022 |
4.384 ± 0.02 | 5.864 ± 0.026 |
6.218 ± 0.048 | 6.455 ± 0.023 |
1.358 ± 0.012 | 4.238 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
