
Hubei sobemo-like virus 10
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.18
Get precalculated fractions of proteins
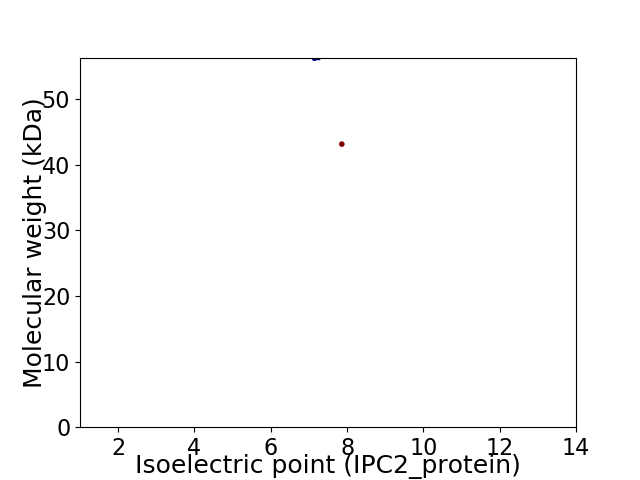
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KET5|A0A1L3KET5_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 10 OX=1923195 PE=4 SV=1
MM1 pKa = 7.11IAKK4 pKa = 9.58VLAFIAWCTKK14 pKa = 7.84GTNLLTLIMALYY26 pKa = 9.88IVNQWLQPVMASMADD41 pKa = 3.81RR42 pKa = 11.84IFEE45 pKa = 4.39VYY47 pKa = 10.21SGLRR51 pKa = 11.84LRR53 pKa = 11.84FNLFWDD59 pKa = 5.42GYY61 pKa = 10.57RR62 pKa = 11.84PGNPGWTFATNTHH75 pKa = 6.11RR76 pKa = 11.84LAYY79 pKa = 9.09QVHH82 pKa = 6.07HH83 pKa = 6.65AWANNMSPPEE93 pKa = 3.67ILGYY97 pKa = 9.17LVCAYY102 pKa = 9.87YY103 pKa = 10.33AAHH106 pKa = 6.09YY107 pKa = 10.21VVILCMFVRR116 pKa = 11.84KK117 pKa = 9.24QLAFLYY123 pKa = 10.66SRR125 pKa = 11.84IQSDD129 pKa = 3.81VLLDD133 pKa = 3.71FAEE136 pKa = 4.41KK137 pKa = 9.48MRR139 pKa = 11.84PGSMLEE145 pKa = 3.93PATNSPKK152 pKa = 10.26FQVEE156 pKa = 3.85VWVRR160 pKa = 11.84TDD162 pKa = 2.64KK163 pKa = 11.17TGFYY167 pKa = 10.54KK168 pKa = 10.52SGQAFCTKK176 pKa = 10.39FGFFTAYY183 pKa = 10.06HH184 pKa = 4.95VVEE187 pKa = 4.86DD188 pKa = 3.83AVEE191 pKa = 4.13VKK193 pKa = 10.37LVNRR197 pKa = 11.84VDD199 pKa = 3.46SQTYY203 pKa = 9.23EE204 pKa = 4.32LIIPASRR211 pKa = 11.84FQQVEE216 pKa = 4.07SDD218 pKa = 3.8VALLTVTSNEE228 pKa = 3.74IGVLQTPQCKK238 pKa = 10.07LMDD241 pKa = 4.95LGLMARR247 pKa = 11.84SGLFVRR253 pKa = 11.84VQAYY257 pKa = 9.67GSQTMGLLEE266 pKa = 4.19PSEE269 pKa = 4.84AFGLCSYY276 pKa = 10.36KK277 pKa = 10.83GSTIKK282 pKa = 10.72GFSGSPYY289 pKa = 8.18YY290 pKa = 10.64CGNNVFGMHH299 pKa = 6.67IGGSSEE305 pKa = 3.88NLGFEE310 pKa = 4.0AAYY313 pKa = 10.45LYY315 pKa = 10.32MLGKK319 pKa = 10.28RR320 pKa = 11.84NSEE323 pKa = 3.94DD324 pKa = 3.59TEE326 pKa = 5.2DD327 pKa = 3.88YY328 pKa = 10.91LLKK331 pKa = 10.54EE332 pKa = 4.34LEE334 pKa = 4.08RR335 pKa = 11.84DD336 pKa = 3.41EE337 pKa = 5.77DD338 pKa = 4.5FVWQRR343 pKa = 11.84SPYY346 pKa = 10.4DD347 pKa = 3.67PDD349 pKa = 3.26EE350 pKa = 4.65ARR352 pKa = 11.84VKK354 pKa = 10.99LGGKK358 pKa = 9.24YY359 pKa = 10.49YY360 pKa = 10.63NVDD363 pKa = 2.69MSTIKK368 pKa = 10.61KK369 pKa = 7.5MEE371 pKa = 3.83KK372 pKa = 9.17MKK374 pKa = 9.95TGRR377 pKa = 11.84RR378 pKa = 11.84APVYY382 pKa = 10.55DD383 pKa = 3.89PDD385 pKa = 4.01YY386 pKa = 10.79EE387 pKa = 4.27EE388 pKa = 5.23EE389 pKa = 4.41GLSEE393 pKa = 4.47ASDD396 pKa = 3.98AEE398 pKa = 4.63TEE400 pKa = 4.26DD401 pKa = 4.12SLPLAPRR408 pKa = 11.84GGLQYY413 pKa = 11.22DD414 pKa = 3.55DD415 pKa = 4.87SKK417 pKa = 12.03NLIAPQQSSAPAGVSGTPTTPQDD440 pKa = 3.74TVPQSALKK448 pKa = 10.01ALNGITLEE456 pKa = 4.06SRR458 pKa = 11.84KK459 pKa = 9.9RR460 pKa = 11.84LAKK463 pKa = 10.45SILDD467 pKa = 3.67GRR469 pKa = 11.84EE470 pKa = 3.76LTPALLRR477 pKa = 11.84DD478 pKa = 3.92LSISTSVSSKK488 pKa = 9.33SKK490 pKa = 10.03RR491 pKa = 11.84RR492 pKa = 11.84SAYY495 pKa = 9.4RR496 pKa = 11.84QKK498 pKa = 11.13KK499 pKa = 9.19KK500 pKa = 10.67NN501 pKa = 3.37
MM1 pKa = 7.11IAKK4 pKa = 9.58VLAFIAWCTKK14 pKa = 7.84GTNLLTLIMALYY26 pKa = 9.88IVNQWLQPVMASMADD41 pKa = 3.81RR42 pKa = 11.84IFEE45 pKa = 4.39VYY47 pKa = 10.21SGLRR51 pKa = 11.84LRR53 pKa = 11.84FNLFWDD59 pKa = 5.42GYY61 pKa = 10.57RR62 pKa = 11.84PGNPGWTFATNTHH75 pKa = 6.11RR76 pKa = 11.84LAYY79 pKa = 9.09QVHH82 pKa = 6.07HH83 pKa = 6.65AWANNMSPPEE93 pKa = 3.67ILGYY97 pKa = 9.17LVCAYY102 pKa = 9.87YY103 pKa = 10.33AAHH106 pKa = 6.09YY107 pKa = 10.21VVILCMFVRR116 pKa = 11.84KK117 pKa = 9.24QLAFLYY123 pKa = 10.66SRR125 pKa = 11.84IQSDD129 pKa = 3.81VLLDD133 pKa = 3.71FAEE136 pKa = 4.41KK137 pKa = 9.48MRR139 pKa = 11.84PGSMLEE145 pKa = 3.93PATNSPKK152 pKa = 10.26FQVEE156 pKa = 3.85VWVRR160 pKa = 11.84TDD162 pKa = 2.64KK163 pKa = 11.17TGFYY167 pKa = 10.54KK168 pKa = 10.52SGQAFCTKK176 pKa = 10.39FGFFTAYY183 pKa = 10.06HH184 pKa = 4.95VVEE187 pKa = 4.86DD188 pKa = 3.83AVEE191 pKa = 4.13VKK193 pKa = 10.37LVNRR197 pKa = 11.84VDD199 pKa = 3.46SQTYY203 pKa = 9.23EE204 pKa = 4.32LIIPASRR211 pKa = 11.84FQQVEE216 pKa = 4.07SDD218 pKa = 3.8VALLTVTSNEE228 pKa = 3.74IGVLQTPQCKK238 pKa = 10.07LMDD241 pKa = 4.95LGLMARR247 pKa = 11.84SGLFVRR253 pKa = 11.84VQAYY257 pKa = 9.67GSQTMGLLEE266 pKa = 4.19PSEE269 pKa = 4.84AFGLCSYY276 pKa = 10.36KK277 pKa = 10.83GSTIKK282 pKa = 10.72GFSGSPYY289 pKa = 8.18YY290 pKa = 10.64CGNNVFGMHH299 pKa = 6.67IGGSSEE305 pKa = 3.88NLGFEE310 pKa = 4.0AAYY313 pKa = 10.45LYY315 pKa = 10.32MLGKK319 pKa = 10.28RR320 pKa = 11.84NSEE323 pKa = 3.94DD324 pKa = 3.59TEE326 pKa = 5.2DD327 pKa = 3.88YY328 pKa = 10.91LLKK331 pKa = 10.54EE332 pKa = 4.34LEE334 pKa = 4.08RR335 pKa = 11.84DD336 pKa = 3.41EE337 pKa = 5.77DD338 pKa = 4.5FVWQRR343 pKa = 11.84SPYY346 pKa = 10.4DD347 pKa = 3.67PDD349 pKa = 3.26EE350 pKa = 4.65ARR352 pKa = 11.84VKK354 pKa = 10.99LGGKK358 pKa = 9.24YY359 pKa = 10.49YY360 pKa = 10.63NVDD363 pKa = 2.69MSTIKK368 pKa = 10.61KK369 pKa = 7.5MEE371 pKa = 3.83KK372 pKa = 9.17MKK374 pKa = 9.95TGRR377 pKa = 11.84RR378 pKa = 11.84APVYY382 pKa = 10.55DD383 pKa = 3.89PDD385 pKa = 4.01YY386 pKa = 10.79EE387 pKa = 4.27EE388 pKa = 5.23EE389 pKa = 4.41GLSEE393 pKa = 4.47ASDD396 pKa = 3.98AEE398 pKa = 4.63TEE400 pKa = 4.26DD401 pKa = 4.12SLPLAPRR408 pKa = 11.84GGLQYY413 pKa = 11.22DD414 pKa = 3.55DD415 pKa = 4.87SKK417 pKa = 12.03NLIAPQQSSAPAGVSGTPTTPQDD440 pKa = 3.74TVPQSALKK448 pKa = 10.01ALNGITLEE456 pKa = 4.06SRR458 pKa = 11.84KK459 pKa = 9.9RR460 pKa = 11.84LAKK463 pKa = 10.45SILDD467 pKa = 3.67GRR469 pKa = 11.84EE470 pKa = 3.76LTPALLRR477 pKa = 11.84DD478 pKa = 3.92LSISTSVSSKK488 pKa = 9.33SKK490 pKa = 10.03RR491 pKa = 11.84RR492 pKa = 11.84SAYY495 pKa = 9.4RR496 pKa = 11.84QKK498 pKa = 11.13KK499 pKa = 9.19KK500 pKa = 10.67NN501 pKa = 3.37
Molecular weight: 56.18 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KET5|A0A1L3KET5_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 10 OX=1923195 PE=4 SV=1
MM1 pKa = 7.72AMTKK5 pKa = 10.63DD6 pKa = 3.15MSRR9 pKa = 11.84YY10 pKa = 7.26WRR12 pKa = 11.84EE13 pKa = 3.92EE14 pKa = 3.19FDD16 pKa = 4.03KK17 pKa = 11.5AMLEE21 pKa = 3.8IDD23 pKa = 3.84MDD25 pKa = 4.43SSPGLCQFAQYY36 pKa = 10.9GPTNAAVFKK45 pKa = 10.69YY46 pKa = 10.42KK47 pKa = 10.78DD48 pKa = 3.79GVWCKK53 pKa = 10.66DD54 pKa = 3.06RR55 pKa = 11.84EE56 pKa = 4.27KK57 pKa = 10.74EE58 pKa = 4.03VRR60 pKa = 11.84DD61 pKa = 3.52VVFFRR66 pKa = 11.84MQQLLRR72 pKa = 11.84GEE74 pKa = 4.52LVADD78 pKa = 4.88DD79 pKa = 3.88IKK81 pKa = 11.04MFVKK85 pKa = 10.54PEE87 pKa = 3.68PHH89 pKa = 6.74KK90 pKa = 10.2PEE92 pKa = 4.42KK93 pKa = 9.29MAQGRR98 pKa = 11.84FRR100 pKa = 11.84LISAVSLVDD109 pKa = 3.6ALVDD113 pKa = 4.72RR114 pKa = 11.84ILFGWLSRR122 pKa = 11.84KK123 pKa = 9.86ALTVVGKK130 pKa = 7.43TPCLTGWSPVHH141 pKa = 6.42GGWKK145 pKa = 9.47YY146 pKa = 9.87IEE148 pKa = 4.51HH149 pKa = 7.0RR150 pKa = 11.84FSGKK154 pKa = 9.53PVVCLDD160 pKa = 3.27KK161 pKa = 11.4SSWDD165 pKa = 3.18WTVQEE170 pKa = 4.34WLITAWFEE178 pKa = 4.11FLKK181 pKa = 11.03GLALGADD188 pKa = 4.01SWWVEE193 pKa = 3.76LVSARR198 pKa = 11.84FQMLFEE204 pKa = 4.19RR205 pKa = 11.84PIYY208 pKa = 10.3QFADD212 pKa = 3.73GTRR215 pKa = 11.84VQQKK219 pKa = 9.36YY220 pKa = 8.91VGIMKK225 pKa = 10.13SGCFLTLILNSVGQSMLHH243 pKa = 5.15YY244 pKa = 9.85VASLRR249 pKa = 11.84IGRR252 pKa = 11.84NPIRR256 pKa = 11.84NQPISVGDD264 pKa = 3.72DD265 pKa = 3.23TTQEE269 pKa = 4.16SFPEE273 pKa = 4.0LEE275 pKa = 4.26EE276 pKa = 4.32YY277 pKa = 10.49INQISRR283 pKa = 11.84LGAKK287 pKa = 9.68VKK289 pKa = 10.25GFKK292 pKa = 9.26VRR294 pKa = 11.84HH295 pKa = 4.27WVEE298 pKa = 4.49FIGFAWIKK306 pKa = 8.94GVCVPAYY313 pKa = 7.02WQKK316 pKa = 11.13HH317 pKa = 4.39LFKK320 pKa = 10.58LQYY323 pKa = 10.6SHH325 pKa = 8.05LEE327 pKa = 4.17DD328 pKa = 3.7VLQSYY333 pKa = 10.11QILYY337 pKa = 10.56ANEE340 pKa = 3.67PVMFEE345 pKa = 4.02LLRR348 pKa = 11.84RR349 pKa = 11.84LAKK352 pKa = 9.85EE353 pKa = 3.97VNPEE357 pKa = 3.85LVMSSYY363 pKa = 9.59EE364 pKa = 3.78AKK366 pKa = 10.74AIMNYY371 pKa = 10.32
MM1 pKa = 7.72AMTKK5 pKa = 10.63DD6 pKa = 3.15MSRR9 pKa = 11.84YY10 pKa = 7.26WRR12 pKa = 11.84EE13 pKa = 3.92EE14 pKa = 3.19FDD16 pKa = 4.03KK17 pKa = 11.5AMLEE21 pKa = 3.8IDD23 pKa = 3.84MDD25 pKa = 4.43SSPGLCQFAQYY36 pKa = 10.9GPTNAAVFKK45 pKa = 10.69YY46 pKa = 10.42KK47 pKa = 10.78DD48 pKa = 3.79GVWCKK53 pKa = 10.66DD54 pKa = 3.06RR55 pKa = 11.84EE56 pKa = 4.27KK57 pKa = 10.74EE58 pKa = 4.03VRR60 pKa = 11.84DD61 pKa = 3.52VVFFRR66 pKa = 11.84MQQLLRR72 pKa = 11.84GEE74 pKa = 4.52LVADD78 pKa = 4.88DD79 pKa = 3.88IKK81 pKa = 11.04MFVKK85 pKa = 10.54PEE87 pKa = 3.68PHH89 pKa = 6.74KK90 pKa = 10.2PEE92 pKa = 4.42KK93 pKa = 9.29MAQGRR98 pKa = 11.84FRR100 pKa = 11.84LISAVSLVDD109 pKa = 3.6ALVDD113 pKa = 4.72RR114 pKa = 11.84ILFGWLSRR122 pKa = 11.84KK123 pKa = 9.86ALTVVGKK130 pKa = 7.43TPCLTGWSPVHH141 pKa = 6.42GGWKK145 pKa = 9.47YY146 pKa = 9.87IEE148 pKa = 4.51HH149 pKa = 7.0RR150 pKa = 11.84FSGKK154 pKa = 9.53PVVCLDD160 pKa = 3.27KK161 pKa = 11.4SSWDD165 pKa = 3.18WTVQEE170 pKa = 4.34WLITAWFEE178 pKa = 4.11FLKK181 pKa = 11.03GLALGADD188 pKa = 4.01SWWVEE193 pKa = 3.76LVSARR198 pKa = 11.84FQMLFEE204 pKa = 4.19RR205 pKa = 11.84PIYY208 pKa = 10.3QFADD212 pKa = 3.73GTRR215 pKa = 11.84VQQKK219 pKa = 9.36YY220 pKa = 8.91VGIMKK225 pKa = 10.13SGCFLTLILNSVGQSMLHH243 pKa = 5.15YY244 pKa = 9.85VASLRR249 pKa = 11.84IGRR252 pKa = 11.84NPIRR256 pKa = 11.84NQPISVGDD264 pKa = 3.72DD265 pKa = 3.23TTQEE269 pKa = 4.16SFPEE273 pKa = 4.0LEE275 pKa = 4.26EE276 pKa = 4.32YY277 pKa = 10.49INQISRR283 pKa = 11.84LGAKK287 pKa = 9.68VKK289 pKa = 10.25GFKK292 pKa = 9.26VRR294 pKa = 11.84HH295 pKa = 4.27WVEE298 pKa = 4.49FIGFAWIKK306 pKa = 8.94GVCVPAYY313 pKa = 7.02WQKK316 pKa = 11.13HH317 pKa = 4.39LFKK320 pKa = 10.58LQYY323 pKa = 10.6SHH325 pKa = 8.05LEE327 pKa = 4.17DD328 pKa = 3.7VLQSYY333 pKa = 10.11QILYY337 pKa = 10.56ANEE340 pKa = 3.67PVMFEE345 pKa = 4.02LLRR348 pKa = 11.84RR349 pKa = 11.84LAKK352 pKa = 9.85EE353 pKa = 3.97VNPEE357 pKa = 3.85LVMSSYY363 pKa = 9.59EE364 pKa = 3.78AKK366 pKa = 10.74AIMNYY371 pKa = 10.32
Molecular weight: 43.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
872 |
371 |
501 |
436.0 |
49.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.11 ± 0.573 | 1.491 ± 0.08 |
4.931 ± 0.05 | 6.078 ± 0.246 |
4.817 ± 0.531 | 6.651 ± 0.284 |
1.491 ± 0.249 | 4.128 ± 0.455 |
6.307 ± 0.441 | 9.862 ± 0.269 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.44 ± 0.21 | 2.982 ± 0.519 |
4.472 ± 0.27 | 4.472 ± 0.239 |
5.505 ± 0.098 | 7.339 ± 0.717 |
4.472 ± 0.948 | 7.454 ± 0.737 |
2.408 ± 0.859 | 4.587 ± 0.512 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
