
Candidatus Jettenia caeni
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Candidatus Brocadiae; Candidatus Brocadiales; Candidatus Brocadiaceae; Candidatus Jettenia
Average proteome isoelectric point is 7.0
Get precalculated fractions of proteins
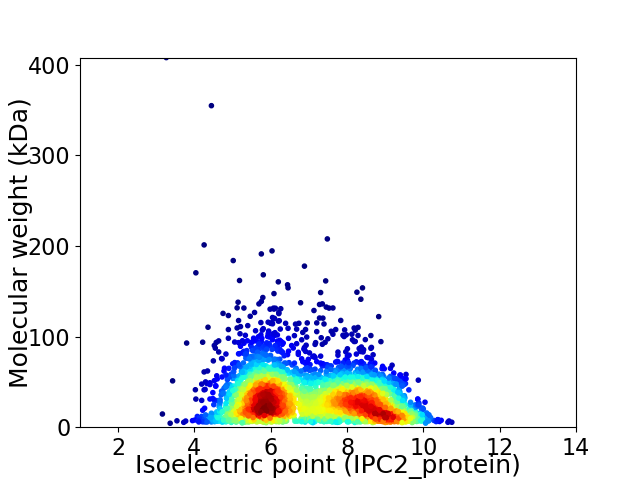
Virtual 2D-PAGE plot for 3545 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I3IM23|I3IM23_9BACT Uncharacterized protein OS=Candidatus Jettenia caeni OX=247490 GN=KSU1_C1172 PE=4 SV=1
MM1 pKa = 7.53LPKK4 pKa = 10.09ILPSVLTMPFAVVLASLLFIVNLATAALPSYY35 pKa = 10.48DD36 pKa = 3.62LQIYY40 pKa = 8.82KK41 pKa = 10.35QGTWYY46 pKa = 9.56TGIAMEE52 pKa = 6.39DD53 pKa = 3.3IDD55 pKa = 4.93NDD57 pKa = 3.76GGLEE61 pKa = 3.93IIIGNRR67 pKa = 11.84NTSSAEE73 pKa = 3.1IWKK76 pKa = 10.35YY77 pKa = 10.64NVSTQTLNQTASISFPYY94 pKa = 9.66HH95 pKa = 5.21IHH97 pKa = 7.63DD98 pKa = 3.65IVAVDD103 pKa = 3.88VDD105 pKa = 3.7HH106 pKa = 7.67DD107 pKa = 4.15GQKK110 pKa = 10.68EE111 pKa = 4.08IIVGIRR117 pKa = 11.84FEE119 pKa = 4.42GLYY122 pKa = 8.88MARR125 pKa = 11.84FSGSAWNVTQIADD138 pKa = 4.11GYY140 pKa = 7.5TWQVLAADD148 pKa = 4.71FNGDD152 pKa = 3.35GHH154 pKa = 8.34VDD156 pKa = 3.03ICQGLDD162 pKa = 2.85HH163 pKa = 7.13AYY165 pKa = 9.78IRR167 pKa = 11.84IFYY170 pKa = 9.57GNGVGGFTLGSSPPLEE186 pKa = 4.03RR187 pKa = 11.84QLGNGWGMNAIDD199 pKa = 4.7LNSDD203 pKa = 2.97GRR205 pKa = 11.84LDD207 pKa = 4.66LIGVVPQWPDD217 pKa = 3.47GIGTGYY223 pKa = 10.48FLRR226 pKa = 11.84AHH228 pKa = 6.99LNLDD232 pKa = 3.27AGGQVTWNSVGPTAPLSYY250 pKa = 10.47DD251 pKa = 3.28QNRR254 pKa = 11.84AQAFLSNSAADD265 pKa = 3.43IDD267 pKa = 4.29GNGYY271 pKa = 9.38IDD273 pKa = 3.47QAVFFANGRR282 pKa = 11.84VSIYY286 pKa = 9.53MGSSSGGQLVWTEE299 pKa = 3.79QQIAQLPTTDD309 pKa = 6.49FIDD312 pKa = 3.58QPSIGISDD320 pKa = 3.77VNNDD324 pKa = 2.87GFLDD328 pKa = 3.53VHH330 pKa = 7.69VDD332 pKa = 3.55GGADD336 pKa = 3.54FNGMVVFLGDD346 pKa = 3.53GTGMYY351 pKa = 8.09QQKK354 pKa = 8.18TLILDD359 pKa = 3.45HH360 pKa = 7.04GIGGFLNSTRR370 pKa = 11.84FGDD373 pKa = 3.72INGDD377 pKa = 3.74GYY379 pKa = 9.69TDD381 pKa = 3.27ICAVRR386 pKa = 11.84FMGNSYY392 pKa = 11.27AGFDD396 pKa = 3.67VLYY399 pKa = 7.85QTPPSNLPPVANAGTDD415 pKa = 3.45QSVHH419 pKa = 5.54VGNTVMLNGSGSSDD433 pKa = 3.53PNGDD437 pKa = 3.71LLTYY441 pKa = 10.43NWTFISKK448 pKa = 9.91PGGSSATLSDD458 pKa = 3.71PTLVNPTFTVDD469 pKa = 2.85KK470 pKa = 10.91AGTYY474 pKa = 9.22KK475 pKa = 10.78VKK477 pKa = 10.79LVVNDD482 pKa = 3.81GTVDD486 pKa = 3.67SVADD490 pKa = 3.64TVLISTINVKK500 pKa = 9.99PVANAGDD507 pKa = 3.9DD508 pKa = 3.37QSVYY512 pKa = 10.76VGSLVTLDD520 pKa = 4.25GSGSSDD526 pKa = 3.41PDD528 pKa = 3.54GDD530 pKa = 3.57QVTYY534 pKa = 10.92NWAFQSKK541 pKa = 8.43PQNSSATLNNPTSASPTFTVDD562 pKa = 2.64KK563 pKa = 10.87AGTYY567 pKa = 7.84VVKK570 pKa = 11.02LIVNDD575 pKa = 4.01GALNSDD581 pKa = 4.18PDD583 pKa = 3.84TVTISNINVAPVADD597 pKa = 4.1AGMDD601 pKa = 3.49QAITTIGTTVQLDD614 pKa = 3.66GTQSYY619 pKa = 11.1DD620 pKa = 3.42PDD622 pKa = 4.69GDD624 pKa = 4.58PITYY628 pKa = 9.21QWAFTSIPDD637 pKa = 3.52GSTASLADD645 pKa = 4.14ADD647 pKa = 4.32TATPTFTADD656 pKa = 2.98VHH658 pKa = 6.98GEE660 pKa = 4.13YY661 pKa = 10.55VVQLVVKK668 pKa = 9.13DD669 pKa = 3.93TSLLSAQDD677 pKa = 3.35AVVISFTNIKK687 pKa = 9.5PVANAGTSQSAVVGDD702 pKa = 3.77TVTINGSGSTDD713 pKa = 3.08GNGDD717 pKa = 3.34NLTYY721 pKa = 10.09QWSFSSVPDD730 pKa = 3.8GSNAEE735 pKa = 4.14IAGPTAITTTFVPDD749 pKa = 3.21KK750 pKa = 10.54AGTYY754 pKa = 9.32VVQLVVNDD762 pKa = 5.58GIVDD766 pKa = 4.0SDD768 pKa = 3.93PSTVQVQVIDD778 pKa = 3.95EE779 pKa = 4.05QTAAIQAIQDD789 pKa = 3.99CEE791 pKa = 3.95QVIASLNSNVFKK803 pKa = 10.59NANMQNTLINKK814 pKa = 9.18LNAVIANIEE823 pKa = 3.97AGNYY827 pKa = 8.86EE828 pKa = 4.28DD829 pKa = 6.22ALGQLRR835 pKa = 11.84NDD837 pKa = 3.58VLGKK841 pKa = 9.1TDD843 pKa = 3.08GCADD847 pKa = 3.6SGSPDD852 pKa = 3.33KK853 pKa = 11.39NDD855 pKa = 3.82WIKK858 pKa = 11.41DD859 pKa = 3.5CAAQGSVYY867 pKa = 10.43PCIQHH872 pKa = 6.86AIVVVEE878 pKa = 3.94GLL880 pKa = 3.55
MM1 pKa = 7.53LPKK4 pKa = 10.09ILPSVLTMPFAVVLASLLFIVNLATAALPSYY35 pKa = 10.48DD36 pKa = 3.62LQIYY40 pKa = 8.82KK41 pKa = 10.35QGTWYY46 pKa = 9.56TGIAMEE52 pKa = 6.39DD53 pKa = 3.3IDD55 pKa = 4.93NDD57 pKa = 3.76GGLEE61 pKa = 3.93IIIGNRR67 pKa = 11.84NTSSAEE73 pKa = 3.1IWKK76 pKa = 10.35YY77 pKa = 10.64NVSTQTLNQTASISFPYY94 pKa = 9.66HH95 pKa = 5.21IHH97 pKa = 7.63DD98 pKa = 3.65IVAVDD103 pKa = 3.88VDD105 pKa = 3.7HH106 pKa = 7.67DD107 pKa = 4.15GQKK110 pKa = 10.68EE111 pKa = 4.08IIVGIRR117 pKa = 11.84FEE119 pKa = 4.42GLYY122 pKa = 8.88MARR125 pKa = 11.84FSGSAWNVTQIADD138 pKa = 4.11GYY140 pKa = 7.5TWQVLAADD148 pKa = 4.71FNGDD152 pKa = 3.35GHH154 pKa = 8.34VDD156 pKa = 3.03ICQGLDD162 pKa = 2.85HH163 pKa = 7.13AYY165 pKa = 9.78IRR167 pKa = 11.84IFYY170 pKa = 9.57GNGVGGFTLGSSPPLEE186 pKa = 4.03RR187 pKa = 11.84QLGNGWGMNAIDD199 pKa = 4.7LNSDD203 pKa = 2.97GRR205 pKa = 11.84LDD207 pKa = 4.66LIGVVPQWPDD217 pKa = 3.47GIGTGYY223 pKa = 10.48FLRR226 pKa = 11.84AHH228 pKa = 6.99LNLDD232 pKa = 3.27AGGQVTWNSVGPTAPLSYY250 pKa = 10.47DD251 pKa = 3.28QNRR254 pKa = 11.84AQAFLSNSAADD265 pKa = 3.43IDD267 pKa = 4.29GNGYY271 pKa = 9.38IDD273 pKa = 3.47QAVFFANGRR282 pKa = 11.84VSIYY286 pKa = 9.53MGSSSGGQLVWTEE299 pKa = 3.79QQIAQLPTTDD309 pKa = 6.49FIDD312 pKa = 3.58QPSIGISDD320 pKa = 3.77VNNDD324 pKa = 2.87GFLDD328 pKa = 3.53VHH330 pKa = 7.69VDD332 pKa = 3.55GGADD336 pKa = 3.54FNGMVVFLGDD346 pKa = 3.53GTGMYY351 pKa = 8.09QQKK354 pKa = 8.18TLILDD359 pKa = 3.45HH360 pKa = 7.04GIGGFLNSTRR370 pKa = 11.84FGDD373 pKa = 3.72INGDD377 pKa = 3.74GYY379 pKa = 9.69TDD381 pKa = 3.27ICAVRR386 pKa = 11.84FMGNSYY392 pKa = 11.27AGFDD396 pKa = 3.67VLYY399 pKa = 7.85QTPPSNLPPVANAGTDD415 pKa = 3.45QSVHH419 pKa = 5.54VGNTVMLNGSGSSDD433 pKa = 3.53PNGDD437 pKa = 3.71LLTYY441 pKa = 10.43NWTFISKK448 pKa = 9.91PGGSSATLSDD458 pKa = 3.71PTLVNPTFTVDD469 pKa = 2.85KK470 pKa = 10.91AGTYY474 pKa = 9.22KK475 pKa = 10.78VKK477 pKa = 10.79LVVNDD482 pKa = 3.81GTVDD486 pKa = 3.67SVADD490 pKa = 3.64TVLISTINVKK500 pKa = 9.99PVANAGDD507 pKa = 3.9DD508 pKa = 3.37QSVYY512 pKa = 10.76VGSLVTLDD520 pKa = 4.25GSGSSDD526 pKa = 3.41PDD528 pKa = 3.54GDD530 pKa = 3.57QVTYY534 pKa = 10.92NWAFQSKK541 pKa = 8.43PQNSSATLNNPTSASPTFTVDD562 pKa = 2.64KK563 pKa = 10.87AGTYY567 pKa = 7.84VVKK570 pKa = 11.02LIVNDD575 pKa = 4.01GALNSDD581 pKa = 4.18PDD583 pKa = 3.84TVTISNINVAPVADD597 pKa = 4.1AGMDD601 pKa = 3.49QAITTIGTTVQLDD614 pKa = 3.66GTQSYY619 pKa = 11.1DD620 pKa = 3.42PDD622 pKa = 4.69GDD624 pKa = 4.58PITYY628 pKa = 9.21QWAFTSIPDD637 pKa = 3.52GSTASLADD645 pKa = 4.14ADD647 pKa = 4.32TATPTFTADD656 pKa = 2.98VHH658 pKa = 6.98GEE660 pKa = 4.13YY661 pKa = 10.55VVQLVVKK668 pKa = 9.13DD669 pKa = 3.93TSLLSAQDD677 pKa = 3.35AVVISFTNIKK687 pKa = 9.5PVANAGTSQSAVVGDD702 pKa = 3.77TVTINGSGSTDD713 pKa = 3.08GNGDD717 pKa = 3.34NLTYY721 pKa = 10.09QWSFSSVPDD730 pKa = 3.8GSNAEE735 pKa = 4.14IAGPTAITTTFVPDD749 pKa = 3.21KK750 pKa = 10.54AGTYY754 pKa = 9.32VVQLVVNDD762 pKa = 5.58GIVDD766 pKa = 4.0SDD768 pKa = 3.93PSTVQVQVIDD778 pKa = 3.95EE779 pKa = 4.05QTAAIQAIQDD789 pKa = 3.99CEE791 pKa = 3.95QVIASLNSNVFKK803 pKa = 10.59NANMQNTLINKK814 pKa = 9.18LNAVIANIEE823 pKa = 3.97AGNYY827 pKa = 8.86EE828 pKa = 4.28DD829 pKa = 6.22ALGQLRR835 pKa = 11.84NDD837 pKa = 3.58VLGKK841 pKa = 9.1TDD843 pKa = 3.08GCADD847 pKa = 3.6SGSPDD852 pKa = 3.33KK853 pKa = 11.39NDD855 pKa = 3.82WIKK858 pKa = 11.41DD859 pKa = 3.5CAAQGSVYY867 pKa = 10.43PCIQHH872 pKa = 6.86AIVVVEE878 pKa = 3.94GLL880 pKa = 3.55
Molecular weight: 92.82 kDa
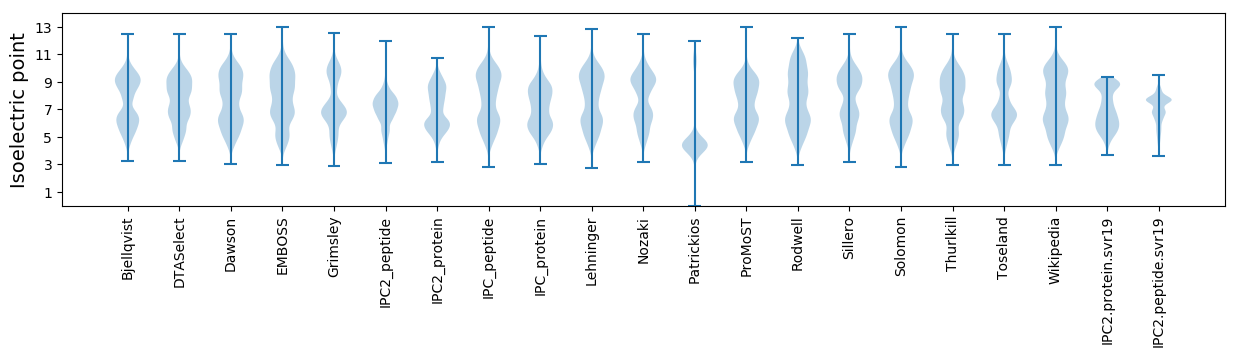
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I3IH07|I3IH07_9BACT Gas vesicle structural protein OS=Candidatus Jettenia caeni OX=247490 GN=KSU1_B0145 PE=3 SV=1
MM1 pKa = 7.42KK2 pKa = 10.72VNIRR6 pKa = 11.84KK7 pKa = 9.66SSIKK11 pKa = 9.61HH12 pKa = 5.21KK13 pKa = 10.38RR14 pKa = 11.84MCGFRR19 pKa = 11.84KK20 pKa = 9.96RR21 pKa = 11.84MRR23 pKa = 11.84TKK25 pKa = 10.35GGRR28 pKa = 11.84AILKK32 pKa = 8.33RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84IGRR39 pKa = 11.84RR40 pKa = 11.84PLLDD44 pKa = 3.21VV45 pKa = 4.01
MM1 pKa = 7.42KK2 pKa = 10.72VNIRR6 pKa = 11.84KK7 pKa = 9.66SSIKK11 pKa = 9.61HH12 pKa = 5.21KK13 pKa = 10.38RR14 pKa = 11.84MCGFRR19 pKa = 11.84KK20 pKa = 9.96RR21 pKa = 11.84MRR23 pKa = 11.84TKK25 pKa = 10.35GGRR28 pKa = 11.84AILKK32 pKa = 8.33RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84IGRR39 pKa = 11.84RR40 pKa = 11.84PLLDD44 pKa = 3.21VV45 pKa = 4.01
Molecular weight: 5.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1108176 |
37 |
3982 |
312.6 |
35.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.453 ± 0.032 | 1.299 ± 0.02 |
5.24 ± 0.039 | 6.824 ± 0.048 |
4.485 ± 0.033 | 6.765 ± 0.043 |
2.241 ± 0.021 | 8.569 ± 0.046 |
7.403 ± 0.049 | 9.294 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.378 ± 0.02 | 4.467 ± 0.028 |
3.958 ± 0.027 | 3.196 ± 0.023 |
4.873 ± 0.032 | 6.095 ± 0.039 |
5.424 ± 0.044 | 6.364 ± 0.036 |
1.073 ± 0.016 | 3.598 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
