
Human parainfluenza virus 4a
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Rubulavirinae; Orthorubulavirus; Human orthorubulavirus 4
Average proteome isoelectric point is 6.72
Get precalculated fractions of proteins
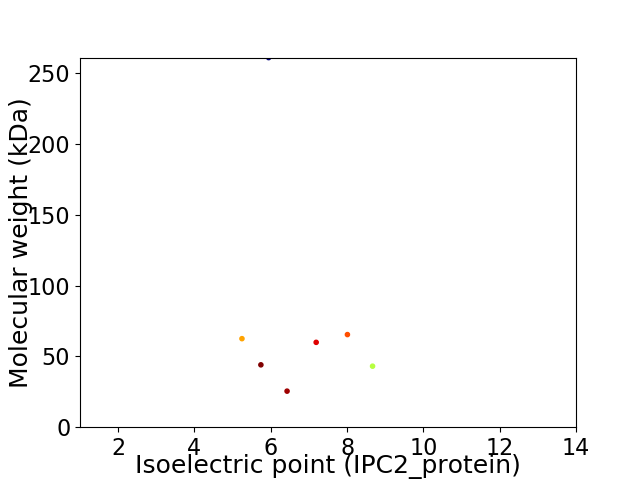
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D7US56|D7US56_9MONO Phosphoprotein OS=Human parainfluenza virus 4a OX=11224 GN=P PE=3 SV=1
MM1 pKa = 7.36SSVLAAYY8 pKa = 7.88EE9 pKa = 4.01QFLQTTEE16 pKa = 3.81DD17 pKa = 3.48RR18 pKa = 11.84GFGDD22 pKa = 3.34QQFVQSDD29 pKa = 3.81TLKK32 pKa = 11.08AEE34 pKa = 3.85IPVFVLNTNDD44 pKa = 3.3PQQRR48 pKa = 11.84FTLMNFCLRR57 pKa = 11.84QAVSSSAKK65 pKa = 10.17SAIKK69 pKa = 10.28QGALLSLLSLQATSMQNHH87 pKa = 6.38LMIAARR93 pKa = 11.84APDD96 pKa = 3.33AALRR100 pKa = 11.84IIEE103 pKa = 4.45VDD105 pKa = 4.13AIDD108 pKa = 4.46PPDD111 pKa = 3.52YY112 pKa = 10.52TLTINPRR119 pKa = 11.84SGWDD123 pKa = 3.61DD124 pKa = 3.06IKK126 pKa = 10.97IRR128 pKa = 11.84AYY130 pKa = 10.26RR131 pKa = 11.84ALSRR135 pKa = 11.84DD136 pKa = 3.71LPISLADD143 pKa = 3.36RR144 pKa = 11.84TVFVSRR150 pKa = 11.84DD151 pKa = 3.38AEE153 pKa = 4.2HH154 pKa = 6.92AVCDD158 pKa = 5.68DD159 pKa = 2.9MDD161 pKa = 3.94TYY163 pKa = 11.5LNRR166 pKa = 11.84IFSVLIQVWIMVCKK180 pKa = 10.62CMTAYY185 pKa = 9.73DD186 pKa = 3.88QPTGSEE192 pKa = 3.93EE193 pKa = 3.9RR194 pKa = 11.84RR195 pKa = 11.84LAKK198 pKa = 10.33YY199 pKa = 9.15KK200 pKa = 9.87QQGRR204 pKa = 11.84MLEE207 pKa = 4.2RR208 pKa = 11.84YY209 pKa = 8.82QLQTDD214 pKa = 3.19ARR216 pKa = 11.84KK217 pKa = 9.88IIQLVIRR224 pKa = 11.84EE225 pKa = 4.33SMVIRR230 pKa = 11.84QFLVQEE236 pKa = 4.35MLTADD241 pKa = 3.31KK242 pKa = 11.11VGAYY246 pKa = 7.06TNRR249 pKa = 11.84YY250 pKa = 7.46YY251 pKa = 11.89AMVGDD256 pKa = 3.78IAKK259 pKa = 10.48YY260 pKa = 8.16IANVGMSAFFLTLKK274 pKa = 10.66FGLGNRR280 pKa = 11.84WKK282 pKa = 9.86PLALAAFSGEE292 pKa = 4.13LVKK295 pKa = 10.99LKK297 pKa = 10.94SLMSLYY303 pKa = 10.24RR304 pKa = 11.84RR305 pKa = 11.84LGDD308 pKa = 3.23RR309 pKa = 11.84SRR311 pKa = 11.84YY312 pKa = 8.99LALLEE317 pKa = 4.48SPEE320 pKa = 4.06LMEE323 pKa = 5.0FAPANYY329 pKa = 9.06PLLFSYY335 pKa = 11.18AMGVGSVQDD344 pKa = 3.78PLIRR348 pKa = 11.84NYY350 pKa = 10.36QFGRR354 pKa = 11.84NFLNTSYY361 pKa = 10.47FQYY364 pKa = 10.83GVEE367 pKa = 4.02TAMKK371 pKa = 9.63HH372 pKa = 5.28QGTVDD377 pKa = 3.31PKK379 pKa = 10.01FASEE383 pKa = 5.07LGITDD388 pKa = 3.81EE389 pKa = 4.55DD390 pKa = 3.97RR391 pKa = 11.84VDD393 pKa = 3.09IMQSVEE399 pKa = 3.63KK400 pKa = 10.16HH401 pKa = 5.22ISGKK405 pKa = 10.66AGDD408 pKa = 5.48DD409 pKa = 3.06ISQPRR414 pKa = 11.84SAFTMSLNRR423 pKa = 11.84SAFITNNNPQDD434 pKa = 3.57LSGARR439 pKa = 11.84LSNYY443 pKa = 7.13EE444 pKa = 4.09QGWSGIDD451 pKa = 3.25QDD453 pKa = 4.19EE454 pKa = 4.52TRR456 pKa = 11.84DD457 pKa = 3.85TLPEE461 pKa = 3.98STMHH465 pKa = 7.05RR466 pKa = 11.84FQNIDD471 pKa = 3.42STNSDD476 pKa = 3.53HH477 pKa = 7.7NEE479 pKa = 3.52LQMPEE484 pKa = 4.26FEE486 pKa = 4.92NDD488 pKa = 2.86INPFNHH494 pKa = 6.44PRR496 pKa = 11.84FTARR500 pKa = 11.84APLIPEE506 pKa = 4.68ISHH509 pKa = 4.62QTPTIRR515 pKa = 11.84MNRR518 pKa = 11.84NVNIRR523 pKa = 11.84DD524 pKa = 3.35STRR527 pKa = 11.84DD528 pKa = 3.33DD529 pKa = 3.5RR530 pKa = 11.84QDD532 pKa = 3.38ANEE535 pKa = 4.65DD536 pKa = 3.45RR537 pKa = 11.84SSNIPDD543 pKa = 5.7DD544 pKa = 3.92ILGDD548 pKa = 3.81LDD550 pKa = 3.62NN551 pKa = 5.04
MM1 pKa = 7.36SSVLAAYY8 pKa = 7.88EE9 pKa = 4.01QFLQTTEE16 pKa = 3.81DD17 pKa = 3.48RR18 pKa = 11.84GFGDD22 pKa = 3.34QQFVQSDD29 pKa = 3.81TLKK32 pKa = 11.08AEE34 pKa = 3.85IPVFVLNTNDD44 pKa = 3.3PQQRR48 pKa = 11.84FTLMNFCLRR57 pKa = 11.84QAVSSSAKK65 pKa = 10.17SAIKK69 pKa = 10.28QGALLSLLSLQATSMQNHH87 pKa = 6.38LMIAARR93 pKa = 11.84APDD96 pKa = 3.33AALRR100 pKa = 11.84IIEE103 pKa = 4.45VDD105 pKa = 4.13AIDD108 pKa = 4.46PPDD111 pKa = 3.52YY112 pKa = 10.52TLTINPRR119 pKa = 11.84SGWDD123 pKa = 3.61DD124 pKa = 3.06IKK126 pKa = 10.97IRR128 pKa = 11.84AYY130 pKa = 10.26RR131 pKa = 11.84ALSRR135 pKa = 11.84DD136 pKa = 3.71LPISLADD143 pKa = 3.36RR144 pKa = 11.84TVFVSRR150 pKa = 11.84DD151 pKa = 3.38AEE153 pKa = 4.2HH154 pKa = 6.92AVCDD158 pKa = 5.68DD159 pKa = 2.9MDD161 pKa = 3.94TYY163 pKa = 11.5LNRR166 pKa = 11.84IFSVLIQVWIMVCKK180 pKa = 10.62CMTAYY185 pKa = 9.73DD186 pKa = 3.88QPTGSEE192 pKa = 3.93EE193 pKa = 3.9RR194 pKa = 11.84RR195 pKa = 11.84LAKK198 pKa = 10.33YY199 pKa = 9.15KK200 pKa = 9.87QQGRR204 pKa = 11.84MLEE207 pKa = 4.2RR208 pKa = 11.84YY209 pKa = 8.82QLQTDD214 pKa = 3.19ARR216 pKa = 11.84KK217 pKa = 9.88IIQLVIRR224 pKa = 11.84EE225 pKa = 4.33SMVIRR230 pKa = 11.84QFLVQEE236 pKa = 4.35MLTADD241 pKa = 3.31KK242 pKa = 11.11VGAYY246 pKa = 7.06TNRR249 pKa = 11.84YY250 pKa = 7.46YY251 pKa = 11.89AMVGDD256 pKa = 3.78IAKK259 pKa = 10.48YY260 pKa = 8.16IANVGMSAFFLTLKK274 pKa = 10.66FGLGNRR280 pKa = 11.84WKK282 pKa = 9.86PLALAAFSGEE292 pKa = 4.13LVKK295 pKa = 10.99LKK297 pKa = 10.94SLMSLYY303 pKa = 10.24RR304 pKa = 11.84RR305 pKa = 11.84LGDD308 pKa = 3.23RR309 pKa = 11.84SRR311 pKa = 11.84YY312 pKa = 8.99LALLEE317 pKa = 4.48SPEE320 pKa = 4.06LMEE323 pKa = 5.0FAPANYY329 pKa = 9.06PLLFSYY335 pKa = 11.18AMGVGSVQDD344 pKa = 3.78PLIRR348 pKa = 11.84NYY350 pKa = 10.36QFGRR354 pKa = 11.84NFLNTSYY361 pKa = 10.47FQYY364 pKa = 10.83GVEE367 pKa = 4.02TAMKK371 pKa = 9.63HH372 pKa = 5.28QGTVDD377 pKa = 3.31PKK379 pKa = 10.01FASEE383 pKa = 5.07LGITDD388 pKa = 3.81EE389 pKa = 4.55DD390 pKa = 3.97RR391 pKa = 11.84VDD393 pKa = 3.09IMQSVEE399 pKa = 3.63KK400 pKa = 10.16HH401 pKa = 5.22ISGKK405 pKa = 10.66AGDD408 pKa = 5.48DD409 pKa = 3.06ISQPRR414 pKa = 11.84SAFTMSLNRR423 pKa = 11.84SAFITNNNPQDD434 pKa = 3.57LSGARR439 pKa = 11.84LSNYY443 pKa = 7.13EE444 pKa = 4.09QGWSGIDD451 pKa = 3.25QDD453 pKa = 4.19EE454 pKa = 4.52TRR456 pKa = 11.84DD457 pKa = 3.85TLPEE461 pKa = 3.98STMHH465 pKa = 7.05RR466 pKa = 11.84FQNIDD471 pKa = 3.42STNSDD476 pKa = 3.53HH477 pKa = 7.7NEE479 pKa = 3.52LQMPEE484 pKa = 4.26FEE486 pKa = 4.92NDD488 pKa = 2.86INPFNHH494 pKa = 6.44PRR496 pKa = 11.84FTARR500 pKa = 11.84APLIPEE506 pKa = 4.68ISHH509 pKa = 4.62QTPTIRR515 pKa = 11.84MNRR518 pKa = 11.84NVNIRR523 pKa = 11.84DD524 pKa = 3.35STRR527 pKa = 11.84DD528 pKa = 3.33DD529 pKa = 3.5RR530 pKa = 11.84QDD532 pKa = 3.38ANEE535 pKa = 4.65DD536 pKa = 3.45RR537 pKa = 11.84SSNIPDD543 pKa = 5.7DD544 pKa = 3.92ILGDD548 pKa = 3.81LDD550 pKa = 3.62NN551 pKa = 5.04
Molecular weight: 62.56 kDa
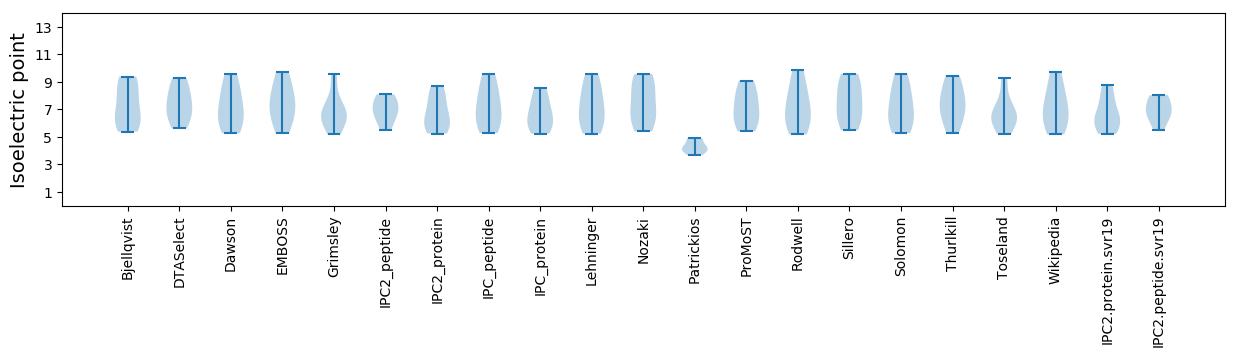
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D7US59|D7US59_9MONO Fusion glycoprotein F0 OS=Human parainfluenza virus 4a OX=11224 GN=F PE=3 SV=1
MM1 pKa = 7.83APTQSKK7 pKa = 9.29VRR9 pKa = 11.84IHH11 pKa = 6.28NLAEE15 pKa = 3.96AHH17 pKa = 6.32EE18 pKa = 4.38KK19 pKa = 9.02TLRR22 pKa = 11.84AFPIEE27 pKa = 4.22VEE29 pKa = 4.19QNPEE33 pKa = 3.92GKK35 pKa = 10.09KK36 pKa = 10.65LLVKK40 pKa = 9.78QIRR43 pKa = 11.84IRR45 pKa = 11.84TLGHH49 pKa = 7.03ADD51 pKa = 3.44HH52 pKa = 7.2SNDD55 pKa = 4.42SICFLNTYY63 pKa = 10.15GFIKK67 pKa = 10.29EE68 pKa = 4.43AVSQTEE74 pKa = 4.0FMRR77 pKa = 11.84AGQKK81 pKa = 9.94SEE83 pKa = 4.21SKK85 pKa = 9.15NTLTACMLPFGPGPNIGSPQKK106 pKa = 9.18MLEE109 pKa = 3.87YY110 pKa = 11.0AEE112 pKa = 5.26DD113 pKa = 3.35IKK115 pKa = 11.28IHH117 pKa = 4.44VRR119 pKa = 11.84KK120 pKa = 7.77TAGCKK125 pKa = 7.59EE126 pKa = 4.1QIVFSLDD133 pKa = 2.84RR134 pKa = 11.84TPQVFRR140 pKa = 11.84GFQFPRR146 pKa = 11.84DD147 pKa = 3.45RR148 pKa = 11.84YY149 pKa = 10.77ACVPSDD155 pKa = 4.69KK156 pKa = 10.65YY157 pKa = 10.9IKK159 pKa = 10.85SPGKK163 pKa = 10.15LVAGPNYY170 pKa = 10.28CYY172 pKa = 10.4IITFLSLTFCPSSQKK187 pKa = 10.46FKK189 pKa = 11.11VPRR192 pKa = 11.84PVLNFRR198 pKa = 11.84STRR201 pKa = 11.84MRR203 pKa = 11.84GIHH206 pKa = 6.45LEE208 pKa = 4.08IIMKK212 pKa = 8.14ITCSEE217 pKa = 3.82NSPIRR222 pKa = 11.84KK223 pKa = 7.65TLITDD228 pKa = 3.67DD229 pKa = 4.8LEE231 pKa = 4.72NGPKK235 pKa = 10.12ASVWIHH241 pKa = 6.15LCNLYY246 pKa = 10.41KK247 pKa = 10.58GRR249 pKa = 11.84NPIKK253 pKa = 10.21IYY255 pKa = 10.85DD256 pKa = 3.67EE257 pKa = 5.13AYY259 pKa = 10.03FAEE262 pKa = 4.88KK263 pKa = 9.48CKK265 pKa = 10.7QMLLSVGISDD275 pKa = 4.1LWGPTIAVHH284 pKa = 6.93ANGKK288 pKa = 8.77IPKK291 pKa = 9.53SASLYY296 pKa = 9.41FNSRR300 pKa = 11.84GWALHH305 pKa = 6.57PIADD309 pKa = 4.8ASPTMAKK316 pKa = 9.53QLWSIGCEE324 pKa = 3.46IMEE327 pKa = 4.62VNAILQGSDD336 pKa = 3.13YY337 pKa = 10.83SALVDD342 pKa = 4.11HH343 pKa = 7.56PDD345 pKa = 3.12VIYY348 pKa = 10.6RR349 pKa = 11.84KK350 pKa = 9.65IRR352 pKa = 11.84IDD354 pKa = 3.53PAKK357 pKa = 10.14KK358 pKa = 10.03QYY360 pKa = 11.02AHH362 pKa = 6.66SKK364 pKa = 8.54WNPFKK369 pKa = 10.84KK370 pKa = 10.31AISMPNLTDD379 pKa = 4.26DD380 pKa = 4.92SII382 pKa = 5.67
MM1 pKa = 7.83APTQSKK7 pKa = 9.29VRR9 pKa = 11.84IHH11 pKa = 6.28NLAEE15 pKa = 3.96AHH17 pKa = 6.32EE18 pKa = 4.38KK19 pKa = 9.02TLRR22 pKa = 11.84AFPIEE27 pKa = 4.22VEE29 pKa = 4.19QNPEE33 pKa = 3.92GKK35 pKa = 10.09KK36 pKa = 10.65LLVKK40 pKa = 9.78QIRR43 pKa = 11.84IRR45 pKa = 11.84TLGHH49 pKa = 7.03ADD51 pKa = 3.44HH52 pKa = 7.2SNDD55 pKa = 4.42SICFLNTYY63 pKa = 10.15GFIKK67 pKa = 10.29EE68 pKa = 4.43AVSQTEE74 pKa = 4.0FMRR77 pKa = 11.84AGQKK81 pKa = 9.94SEE83 pKa = 4.21SKK85 pKa = 9.15NTLTACMLPFGPGPNIGSPQKK106 pKa = 9.18MLEE109 pKa = 3.87YY110 pKa = 11.0AEE112 pKa = 5.26DD113 pKa = 3.35IKK115 pKa = 11.28IHH117 pKa = 4.44VRR119 pKa = 11.84KK120 pKa = 7.77TAGCKK125 pKa = 7.59EE126 pKa = 4.1QIVFSLDD133 pKa = 2.84RR134 pKa = 11.84TPQVFRR140 pKa = 11.84GFQFPRR146 pKa = 11.84DD147 pKa = 3.45RR148 pKa = 11.84YY149 pKa = 10.77ACVPSDD155 pKa = 4.69KK156 pKa = 10.65YY157 pKa = 10.9IKK159 pKa = 10.85SPGKK163 pKa = 10.15LVAGPNYY170 pKa = 10.28CYY172 pKa = 10.4IITFLSLTFCPSSQKK187 pKa = 10.46FKK189 pKa = 11.11VPRR192 pKa = 11.84PVLNFRR198 pKa = 11.84STRR201 pKa = 11.84MRR203 pKa = 11.84GIHH206 pKa = 6.45LEE208 pKa = 4.08IIMKK212 pKa = 8.14ITCSEE217 pKa = 3.82NSPIRR222 pKa = 11.84KK223 pKa = 7.65TLITDD228 pKa = 3.67DD229 pKa = 4.8LEE231 pKa = 4.72NGPKK235 pKa = 10.12ASVWIHH241 pKa = 6.15LCNLYY246 pKa = 10.41KK247 pKa = 10.58GRR249 pKa = 11.84NPIKK253 pKa = 10.21IYY255 pKa = 10.85DD256 pKa = 3.67EE257 pKa = 5.13AYY259 pKa = 10.03FAEE262 pKa = 4.88KK263 pKa = 9.48CKK265 pKa = 10.7QMLLSVGISDD275 pKa = 4.1LWGPTIAVHH284 pKa = 6.93ANGKK288 pKa = 8.77IPKK291 pKa = 9.53SASLYY296 pKa = 9.41FNSRR300 pKa = 11.84GWALHH305 pKa = 6.57PIADD309 pKa = 4.8ASPTMAKK316 pKa = 9.53QLWSIGCEE324 pKa = 3.46IMEE327 pKa = 4.62VNAILQGSDD336 pKa = 3.13YY337 pKa = 10.83SALVDD342 pKa = 4.11HH343 pKa = 7.56PDD345 pKa = 3.12VIYY348 pKa = 10.6RR349 pKa = 11.84KK350 pKa = 9.65IRR352 pKa = 11.84IDD354 pKa = 3.53PAKK357 pKa = 10.14KK358 pKa = 10.03QYY360 pKa = 11.02AHH362 pKa = 6.66SKK364 pKa = 8.54WNPFKK369 pKa = 10.84KK370 pKa = 10.31AISMPNLTDD379 pKa = 4.26DD380 pKa = 4.92SII382 pKa = 5.67
Molecular weight: 43.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4957 |
229 |
2279 |
708.1 |
80.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.185 ± 0.63 | 2.038 ± 0.329 |
5.487 ± 0.479 | 5.144 ± 0.51 |
3.651 ± 0.564 | 4.6 ± 0.442 |
2.098 ± 0.262 | 8.856 ± 0.747 |
5.709 ± 0.427 | 10.874 ± 0.945 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.098 ± 0.255 | 6.617 ± 0.284 |
4.741 ± 0.367 | 4.66 ± 0.385 |
4.882 ± 0.455 | 7.888 ± 0.308 |
6.415 ± 0.709 | 4.438 ± 0.307 |
1.089 ± 0.275 | 3.53 ± 0.511 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
