
Rice yellow mottle virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Sobemovirus
Average proteome isoelectric point is 6.73
Get precalculated fractions of proteins
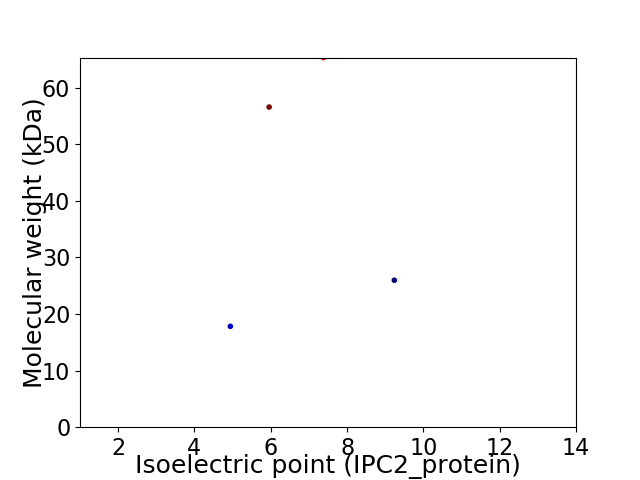
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
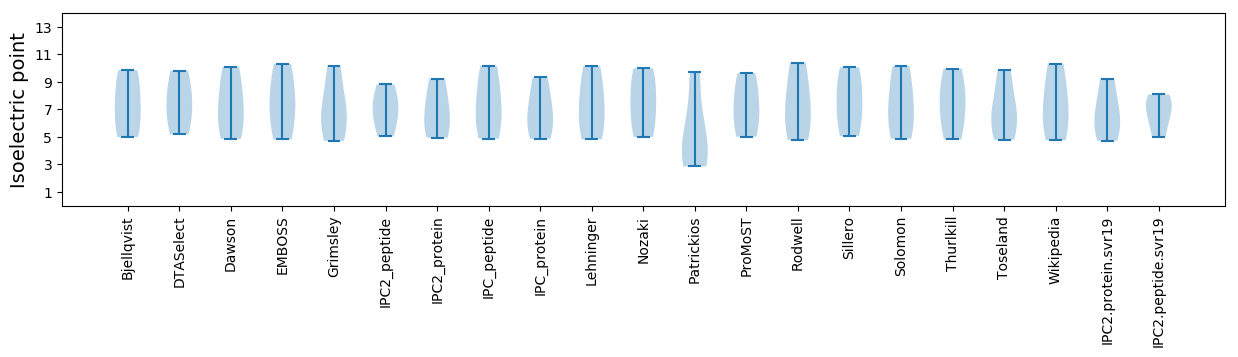
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9DH71|Q9DH71_9VIRU Capsid protein OS=Rice yellow mottle virus OX=31744 GN=CP PE=2 SV=1
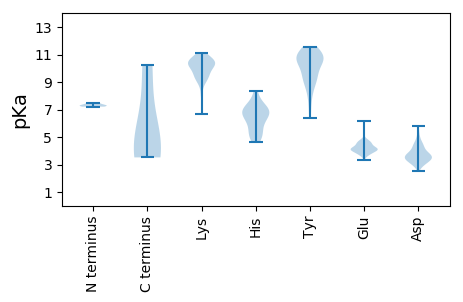
MM1 pKa = 7.21TRR3 pKa = 11.84LEE5 pKa = 4.02VLIRR9 pKa = 11.84PTQQTVAKK17 pKa = 9.05AIAVGYY23 pKa = 6.42THH25 pKa = 7.55ALTWVWHH32 pKa = 4.94SQTWDD37 pKa = 2.89VDD39 pKa = 3.92AVSDD43 pKa = 4.2PVLSADD49 pKa = 4.14FNPEE53 pKa = 3.3KK54 pKa = 10.81VGWVSVSFACTRR66 pKa = 11.84CTAHH70 pKa = 6.92YY71 pKa = 7.3YY72 pKa = 9.93TSEE75 pKa = 3.88QVKK78 pKa = 9.43YY79 pKa = 9.57FVNIPPVHH87 pKa = 6.52YY88 pKa = 10.56DD89 pKa = 3.55VVCADD94 pKa = 4.57CEE96 pKa = 4.13RR97 pKa = 11.84SVQQDD102 pKa = 3.48DD103 pKa = 4.48EE104 pKa = 4.7IDD106 pKa = 3.74RR107 pKa = 11.84EE108 pKa = 4.06HH109 pKa = 7.61DD110 pKa = 3.29EE111 pKa = 4.3RR112 pKa = 11.84NAEE115 pKa = 3.79ISACNARR122 pKa = 11.84ALSEE126 pKa = 4.13GRR128 pKa = 11.84PASLVYY134 pKa = 10.44LSRR137 pKa = 11.84DD138 pKa = 3.07ACDD141 pKa = 3.44IPEE144 pKa = 4.24HH145 pKa = 6.76SGTCRR150 pKa = 11.84FDD152 pKa = 3.18KK153 pKa = 10.97YY154 pKa = 11.58LNFF157 pKa = 4.56
MM1 pKa = 7.21TRR3 pKa = 11.84LEE5 pKa = 4.02VLIRR9 pKa = 11.84PTQQTVAKK17 pKa = 9.05AIAVGYY23 pKa = 6.42THH25 pKa = 7.55ALTWVWHH32 pKa = 4.94SQTWDD37 pKa = 2.89VDD39 pKa = 3.92AVSDD43 pKa = 4.2PVLSADD49 pKa = 4.14FNPEE53 pKa = 3.3KK54 pKa = 10.81VGWVSVSFACTRR66 pKa = 11.84CTAHH70 pKa = 6.92YY71 pKa = 7.3YY72 pKa = 9.93TSEE75 pKa = 3.88QVKK78 pKa = 9.43YY79 pKa = 9.57FVNIPPVHH87 pKa = 6.52YY88 pKa = 10.56DD89 pKa = 3.55VVCADD94 pKa = 4.57CEE96 pKa = 4.13RR97 pKa = 11.84SVQQDD102 pKa = 3.48DD103 pKa = 4.48EE104 pKa = 4.7IDD106 pKa = 3.74RR107 pKa = 11.84EE108 pKa = 4.06HH109 pKa = 7.61DD110 pKa = 3.29EE111 pKa = 4.3RR112 pKa = 11.84NAEE115 pKa = 3.79ISACNARR122 pKa = 11.84ALSEE126 pKa = 4.13GRR128 pKa = 11.84PASLVYY134 pKa = 10.44LSRR137 pKa = 11.84DD138 pKa = 3.07ACDD141 pKa = 3.44IPEE144 pKa = 4.24HH145 pKa = 6.76SGTCRR150 pKa = 11.84FDD152 pKa = 3.18KK153 pKa = 10.97YY154 pKa = 11.58LNFF157 pKa = 4.56
Molecular weight: 17.82 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9DH71|Q9DH71_9VIRU Capsid protein OS=Rice yellow mottle virus OX=31744 GN=CP PE=2 SV=1
MM1 pKa = 7.29ARR3 pKa = 11.84KK4 pKa = 9.21GKK6 pKa = 8.36KK7 pKa = 7.99TNSNQGQQGKK17 pKa = 9.92RR18 pKa = 11.84KK19 pKa = 8.91GRR21 pKa = 11.84RR22 pKa = 11.84PRR24 pKa = 11.84GRR26 pKa = 11.84SAEE29 pKa = 4.1PQLQRR34 pKa = 11.84APVAQASRR42 pKa = 11.84ISGTVPGPLSSNTWPLHH59 pKa = 4.61SVEE62 pKa = 4.68FLADD66 pKa = 4.59FKK68 pKa = 11.12RR69 pKa = 11.84SSTSADD75 pKa = 2.85ATTYY79 pKa = 11.22DD80 pKa = 4.09CVPFNLPRR88 pKa = 11.84VWSLARR94 pKa = 11.84CYY96 pKa = 11.33SMWKK100 pKa = 6.65PTRR103 pKa = 11.84WDD105 pKa = 3.27VVYY108 pKa = 10.44LPEE111 pKa = 4.59VSATVAGSIEE121 pKa = 4.04MCFLYY126 pKa = 10.32DD127 pKa = 3.68YY128 pKa = 11.33ADD130 pKa = 4.86TIPSDD135 pKa = 3.41TGKK138 pKa = 9.75MSRR141 pKa = 11.84TAGFVTSSVWYY152 pKa = 9.41GAEE155 pKa = 4.21GCHH158 pKa = 6.29LLSGGSARR166 pKa = 11.84NAVVASMDD174 pKa = 3.68CSRR177 pKa = 11.84VGWKK181 pKa = 9.93RR182 pKa = 11.84VTSSIPSSVDD192 pKa = 3.03PNVVNTILPARR203 pKa = 11.84LAVRR207 pKa = 11.84SSIKK211 pKa = 9.37PTVSDD216 pKa = 3.84TPGKK220 pKa = 10.24LYY222 pKa = 11.0VIASMVLRR230 pKa = 11.84DD231 pKa = 4.13PVDD234 pKa = 3.43PTLNTT239 pKa = 3.52
MM1 pKa = 7.29ARR3 pKa = 11.84KK4 pKa = 9.21GKK6 pKa = 8.36KK7 pKa = 7.99TNSNQGQQGKK17 pKa = 9.92RR18 pKa = 11.84KK19 pKa = 8.91GRR21 pKa = 11.84RR22 pKa = 11.84PRR24 pKa = 11.84GRR26 pKa = 11.84SAEE29 pKa = 4.1PQLQRR34 pKa = 11.84APVAQASRR42 pKa = 11.84ISGTVPGPLSSNTWPLHH59 pKa = 4.61SVEE62 pKa = 4.68FLADD66 pKa = 4.59FKK68 pKa = 11.12RR69 pKa = 11.84SSTSADD75 pKa = 2.85ATTYY79 pKa = 11.22DD80 pKa = 4.09CVPFNLPRR88 pKa = 11.84VWSLARR94 pKa = 11.84CYY96 pKa = 11.33SMWKK100 pKa = 6.65PTRR103 pKa = 11.84WDD105 pKa = 3.27VVYY108 pKa = 10.44LPEE111 pKa = 4.59VSATVAGSIEE121 pKa = 4.04MCFLYY126 pKa = 10.32DD127 pKa = 3.68YY128 pKa = 11.33ADD130 pKa = 4.86TIPSDD135 pKa = 3.41TGKK138 pKa = 9.75MSRR141 pKa = 11.84TAGFVTSSVWYY152 pKa = 9.41GAEE155 pKa = 4.21GCHH158 pKa = 6.29LLSGGSARR166 pKa = 11.84NAVVASMDD174 pKa = 3.68CSRR177 pKa = 11.84VGWKK181 pKa = 9.93RR182 pKa = 11.84VTSSIPSSVDD192 pKa = 3.03PNVVNTILPARR203 pKa = 11.84LAVRR207 pKa = 11.84SSIKK211 pKa = 9.37PTVSDD216 pKa = 3.84TPGKK220 pKa = 10.24LYY222 pKa = 11.0VIASMVLRR230 pKa = 11.84DD231 pKa = 4.13PVDD234 pKa = 3.43PTLNTT239 pKa = 3.52
Molecular weight: 25.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1505 |
157 |
605 |
376.3 |
41.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.508 ± 0.504 | 2.326 ± 0.578 |
5.05 ± 0.595 | 5.847 ± 0.984 |
3.256 ± 0.424 | 7.375 ± 0.91 |
2.458 ± 0.431 | 4.12 ± 0.318 |
4.186 ± 0.466 | 7.508 ± 0.789 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.329 ± 0.292 | 2.99 ± 0.173 |
6.246 ± 0.502 | 3.256 ± 0.438 |
6.91 ± 0.589 | 11.096 ± 1.381 |
5.449 ± 0.837 | 8.173 ± 0.882 |
2.259 ± 0.576 | 2.658 ± 0.407 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
