
Eremococcus coleocola ACS-139-V-Col8
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Aerococcaceae; Eremococcus; Eremococcus coleocola
Average proteome isoelectric point is 5.97
Get precalculated fractions of proteins
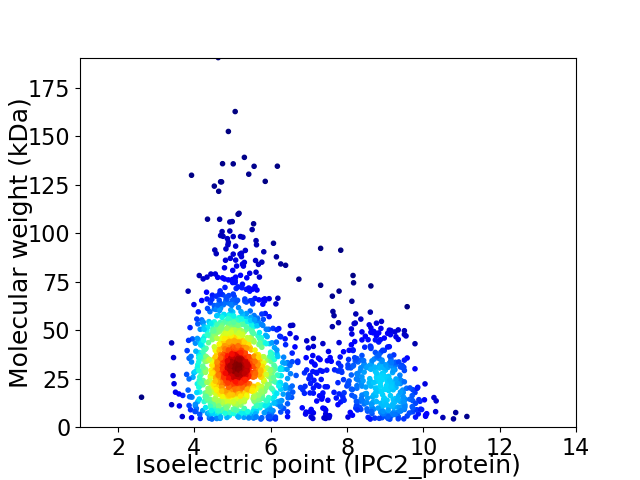
Virtual 2D-PAGE plot for 1720 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E4KQN8|E4KQN8_9LACT Oxaloacetate decarboxylase gamma chain OS=Eremococcus coleocola ACS-139-V-Col8 OX=908337 GN=HMPREF9257_0597 PE=3 SV=1
MM1 pKa = 7.23KK2 pKa = 10.42NKK4 pKa = 9.93YY5 pKa = 10.1SKK7 pKa = 10.64LSKK10 pKa = 9.18TALVLMSSLVLQAPLALAQEE30 pKa = 5.2AYY32 pKa = 9.06TVQQGDD38 pKa = 3.78TLYY41 pKa = 11.31DD42 pKa = 3.82LAQTIGASVDD52 pKa = 4.12EE53 pKa = 4.12LMAWNGLTSDD63 pKa = 4.11YY64 pKa = 11.11LDD66 pKa = 3.9VGDD69 pKa = 5.05ILYY72 pKa = 9.25LAPAAPSAEE81 pKa = 4.32STPEE85 pKa = 4.03TTSPAYY91 pKa = 10.85SNGPGYY97 pKa = 8.8HH98 pKa = 5.38TVSAGEE104 pKa = 4.05NLWSIAYY111 pKa = 10.16YY112 pKa = 10.75YY113 pKa = 8.34GTTMQAIVDD122 pKa = 4.04ANGLSSDD129 pKa = 3.89YY130 pKa = 11.18LDD132 pKa = 5.14VGDD135 pKa = 4.35QLYY138 pKa = 10.39IPAVNDD144 pKa = 3.6AYY146 pKa = 10.83GPGEE150 pKa = 3.84AVSYY154 pKa = 10.28YY155 pKa = 10.71GQEE158 pKa = 3.66QSTNSTPADD167 pKa = 3.6YY168 pKa = 10.7SSSVTPDD175 pKa = 3.0STVKK179 pKa = 10.76DD180 pKa = 3.72GAQIHH185 pKa = 5.35TVVAGDD191 pKa = 3.77NLYY194 pKa = 10.79DD195 pKa = 3.3IAIAYY200 pKa = 7.64GINLNDD206 pKa = 4.0LYY208 pKa = 10.35TWNGLSSDD216 pKa = 3.99YY217 pKa = 11.4LNVGDD222 pKa = 4.19QLIVGLAGPVEE233 pKa = 4.63DD234 pKa = 3.98NQEE237 pKa = 3.9VTVTDD242 pKa = 4.09DD243 pKa = 2.94QGQSKK248 pKa = 10.62KK249 pKa = 9.88IDD251 pKa = 3.45LSKK254 pKa = 10.51IPEE257 pKa = 4.02KK258 pKa = 10.49FRR260 pKa = 11.84PKK262 pKa = 9.96TYY264 pKa = 9.98TVVAGDD270 pKa = 4.26NVWKK274 pKa = 10.3IAQKK278 pKa = 10.72NKK280 pKa = 9.75VSAEE284 pKa = 4.18SIRR287 pKa = 11.84LWNDD291 pKa = 2.84LAEE294 pKa = 4.6DD295 pKa = 3.36QGLTVGDD302 pKa = 3.54QVYY305 pKa = 10.17VSNPALVPEE314 pKa = 3.87IHH316 pKa = 6.37EE317 pKa = 4.36VVEE320 pKa = 4.64GEE322 pKa = 4.17DD323 pKa = 3.57LASIGADD330 pKa = 3.33YY331 pKa = 10.73QVAEE335 pKa = 5.0DD336 pKa = 6.28DD337 pKa = 3.97LLAWNDD343 pKa = 2.83ISKK346 pKa = 11.27AEE348 pKa = 4.61DD349 pKa = 3.24IKK351 pKa = 11.55VGDD354 pKa = 4.26QIAVSNPKK362 pKa = 9.58PEE364 pKa = 3.88KK365 pKa = 9.99HH366 pKa = 4.97QVQAGEE372 pKa = 4.14TLEE375 pKa = 5.92DD376 pKa = 3.07IANTYY381 pKa = 9.82GVTVDD386 pKa = 6.36DD387 pKa = 4.45LRR389 pKa = 11.84TWNNLPANALIVNGEE404 pKa = 4.7LIVADD409 pKa = 4.03PQAGNSAEE417 pKa = 3.96
MM1 pKa = 7.23KK2 pKa = 10.42NKK4 pKa = 9.93YY5 pKa = 10.1SKK7 pKa = 10.64LSKK10 pKa = 9.18TALVLMSSLVLQAPLALAQEE30 pKa = 5.2AYY32 pKa = 9.06TVQQGDD38 pKa = 3.78TLYY41 pKa = 11.31DD42 pKa = 3.82LAQTIGASVDD52 pKa = 4.12EE53 pKa = 4.12LMAWNGLTSDD63 pKa = 4.11YY64 pKa = 11.11LDD66 pKa = 3.9VGDD69 pKa = 5.05ILYY72 pKa = 9.25LAPAAPSAEE81 pKa = 4.32STPEE85 pKa = 4.03TTSPAYY91 pKa = 10.85SNGPGYY97 pKa = 8.8HH98 pKa = 5.38TVSAGEE104 pKa = 4.05NLWSIAYY111 pKa = 10.16YY112 pKa = 10.75YY113 pKa = 8.34GTTMQAIVDD122 pKa = 4.04ANGLSSDD129 pKa = 3.89YY130 pKa = 11.18LDD132 pKa = 5.14VGDD135 pKa = 4.35QLYY138 pKa = 10.39IPAVNDD144 pKa = 3.6AYY146 pKa = 10.83GPGEE150 pKa = 3.84AVSYY154 pKa = 10.28YY155 pKa = 10.71GQEE158 pKa = 3.66QSTNSTPADD167 pKa = 3.6YY168 pKa = 10.7SSSVTPDD175 pKa = 3.0STVKK179 pKa = 10.76DD180 pKa = 3.72GAQIHH185 pKa = 5.35TVVAGDD191 pKa = 3.77NLYY194 pKa = 10.79DD195 pKa = 3.3IAIAYY200 pKa = 7.64GINLNDD206 pKa = 4.0LYY208 pKa = 10.35TWNGLSSDD216 pKa = 3.99YY217 pKa = 11.4LNVGDD222 pKa = 4.19QLIVGLAGPVEE233 pKa = 4.63DD234 pKa = 3.98NQEE237 pKa = 3.9VTVTDD242 pKa = 4.09DD243 pKa = 2.94QGQSKK248 pKa = 10.62KK249 pKa = 9.88IDD251 pKa = 3.45LSKK254 pKa = 10.51IPEE257 pKa = 4.02KK258 pKa = 10.49FRR260 pKa = 11.84PKK262 pKa = 9.96TYY264 pKa = 9.98TVVAGDD270 pKa = 4.26NVWKK274 pKa = 10.3IAQKK278 pKa = 10.72NKK280 pKa = 9.75VSAEE284 pKa = 4.18SIRR287 pKa = 11.84LWNDD291 pKa = 2.84LAEE294 pKa = 4.6DD295 pKa = 3.36QGLTVGDD302 pKa = 3.54QVYY305 pKa = 10.17VSNPALVPEE314 pKa = 3.87IHH316 pKa = 6.37EE317 pKa = 4.36VVEE320 pKa = 4.64GEE322 pKa = 4.17DD323 pKa = 3.57LASIGADD330 pKa = 3.33YY331 pKa = 10.73QVAEE335 pKa = 5.0DD336 pKa = 6.28DD337 pKa = 3.97LLAWNDD343 pKa = 2.83ISKK346 pKa = 11.27AEE348 pKa = 4.61DD349 pKa = 3.24IKK351 pKa = 11.55VGDD354 pKa = 4.26QIAVSNPKK362 pKa = 9.58PEE364 pKa = 3.88KK365 pKa = 9.99HH366 pKa = 4.97QVQAGEE372 pKa = 4.14TLEE375 pKa = 5.92DD376 pKa = 3.07IANTYY381 pKa = 9.82GVTVDD386 pKa = 6.36DD387 pKa = 4.45LRR389 pKa = 11.84TWNNLPANALIVNGEE404 pKa = 4.7LIVADD409 pKa = 4.03PQAGNSAEE417 pKa = 3.96
Molecular weight: 44.73 kDa
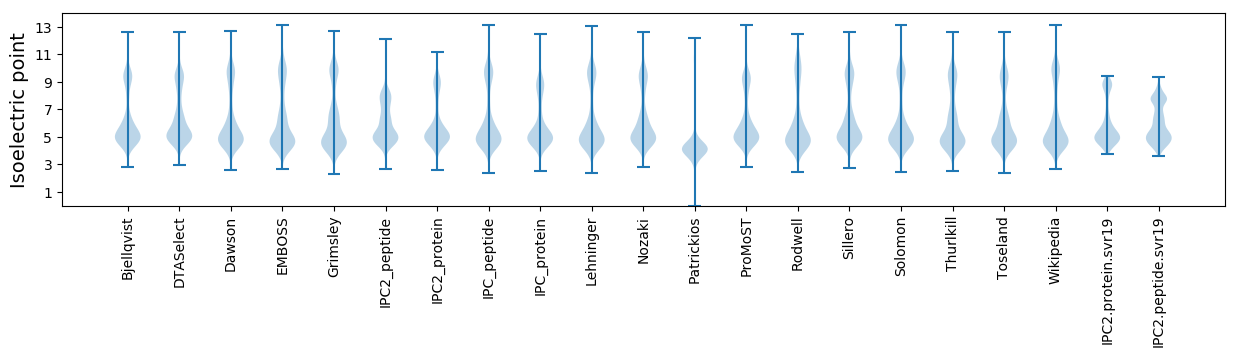
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E4KR82|E4KR82_9LACT Uncharacterized protein OS=Eremococcus coleocola ACS-139-V-Col8 OX=908337 GN=HMPREF9257_0381 PE=4 SV=1
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.69QPKK8 pKa = 9.2KK9 pKa = 7.96RR10 pKa = 11.84HH11 pKa = 4.8RR12 pKa = 11.84QKK14 pKa = 10.52VHH16 pKa = 5.94GFRR19 pKa = 11.84QRR21 pKa = 11.84MKK23 pKa = 8.52TKK25 pKa = 9.67NGRR28 pKa = 11.84RR29 pKa = 11.84VLRR32 pKa = 11.84KK33 pKa = 9.21RR34 pKa = 11.84RR35 pKa = 11.84QKK37 pKa = 10.45GRR39 pKa = 11.84KK40 pKa = 8.5SLAVV44 pKa = 3.3
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.69QPKK8 pKa = 9.2KK9 pKa = 7.96RR10 pKa = 11.84HH11 pKa = 4.8RR12 pKa = 11.84QKK14 pKa = 10.52VHH16 pKa = 5.94GFRR19 pKa = 11.84QRR21 pKa = 11.84MKK23 pKa = 8.52TKK25 pKa = 9.67NGRR28 pKa = 11.84RR29 pKa = 11.84VLRR32 pKa = 11.84KK33 pKa = 9.21RR34 pKa = 11.84RR35 pKa = 11.84QKK37 pKa = 10.45GRR39 pKa = 11.84KK40 pKa = 8.5SLAVV44 pKa = 3.3
Molecular weight: 5.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
512594 |
37 |
1721 |
298.0 |
33.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.933 ± 0.061 | 0.532 ± 0.015 |
6.217 ± 0.049 | 6.485 ± 0.061 |
4.285 ± 0.041 | 6.526 ± 0.055 |
1.986 ± 0.029 | 7.492 ± 0.05 |
6.05 ± 0.049 | 10.249 ± 0.074 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.666 ± 0.028 | 4.505 ± 0.043 |
3.517 ± 0.036 | 5.237 ± 0.063 |
3.849 ± 0.045 | 5.947 ± 0.04 |
5.058 ± 0.04 | 6.735 ± 0.052 |
0.882 ± 0.021 | 3.847 ± 0.042 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
