
Phytophthora parasitica virus
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 5.89
Get precalculated fractions of proteins
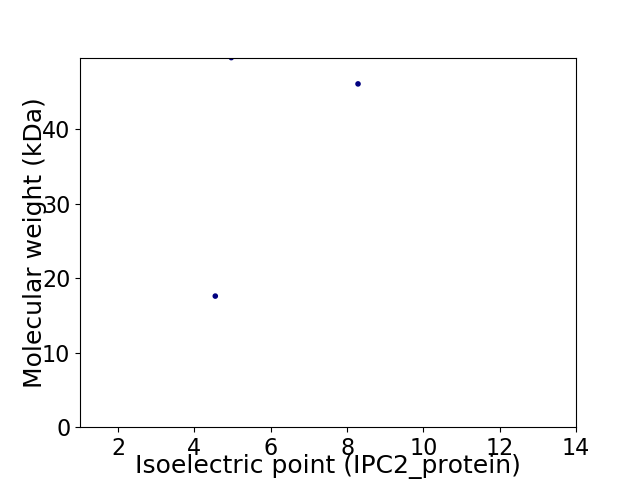
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
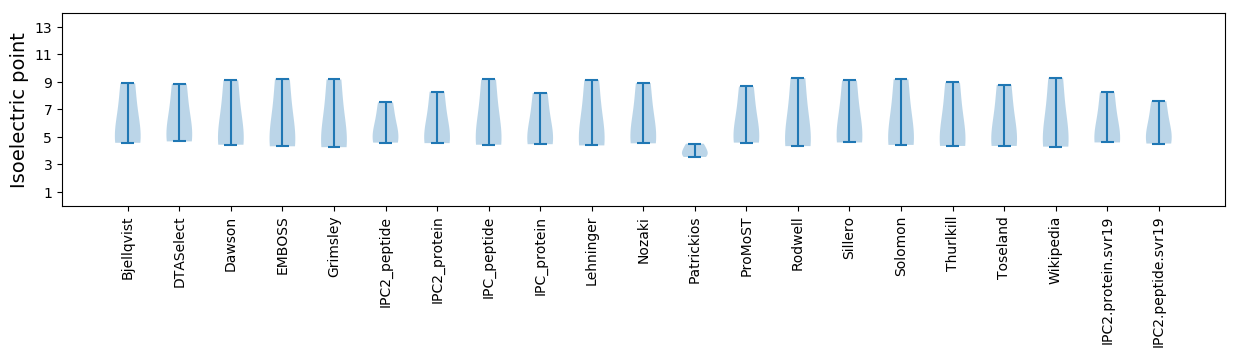
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C5AP07|A0A0C5AP07_9VIRU Isolate 1 ORF3 putative replication protein and putative capsid protein genes OS=Phytophthora parasitica virus OX=1608451 PE=4 SV=1
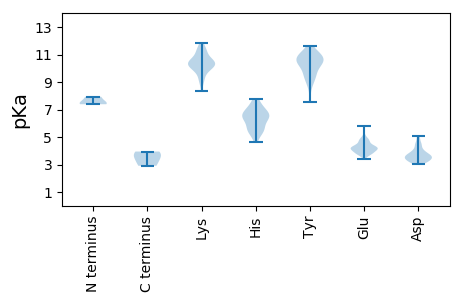
MM1 pKa = 7.47AHH3 pKa = 6.02EE4 pKa = 4.01QHH6 pKa = 6.57HH7 pKa = 6.61NIVTEE12 pKa = 4.07LVEE15 pKa = 4.22SFIVEE20 pKa = 4.13EE21 pKa = 4.26NKK23 pKa = 10.52ALHH26 pKa = 6.73DD27 pKa = 4.12KK28 pKa = 10.94LRR30 pKa = 11.84TALNEE35 pKa = 3.88AQALRR40 pKa = 11.84LRR42 pKa = 11.84LTRR45 pKa = 11.84LNTLYY50 pKa = 11.52NMAGGLNGRR59 pKa = 11.84MQQEE63 pKa = 4.13LRR65 pKa = 11.84DD66 pKa = 3.54LHH68 pKa = 6.82AAIEE72 pKa = 4.16MMDD75 pKa = 4.39LIARR79 pKa = 11.84EE80 pKa = 3.92YY81 pKa = 10.64FINNAEE87 pKa = 3.96ARR89 pKa = 11.84ARR91 pKa = 11.84WAPYY95 pKa = 9.88CAFEE99 pKa = 5.81DD100 pKa = 5.06DD101 pKa = 4.54EE102 pKa = 4.48FHH104 pKa = 7.49HH105 pKa = 7.2LIAEE109 pKa = 4.42ANAGFPSDD117 pKa = 3.82LAIVMEE123 pKa = 4.34PTQEE127 pKa = 4.78DD128 pKa = 3.71EE129 pKa = 4.45DD130 pKa = 4.99DD131 pKa = 4.02RR132 pKa = 11.84DD133 pKa = 3.84SVIDD137 pKa = 3.6ILEE140 pKa = 4.25LTEE143 pKa = 4.68KK144 pKa = 10.54EE145 pKa = 4.17MRR147 pKa = 11.84DD148 pKa = 3.76FMDD151 pKa = 3.13VV152 pKa = 2.91
MM1 pKa = 7.47AHH3 pKa = 6.02EE4 pKa = 4.01QHH6 pKa = 6.57HH7 pKa = 6.61NIVTEE12 pKa = 4.07LVEE15 pKa = 4.22SFIVEE20 pKa = 4.13EE21 pKa = 4.26NKK23 pKa = 10.52ALHH26 pKa = 6.73DD27 pKa = 4.12KK28 pKa = 10.94LRR30 pKa = 11.84TALNEE35 pKa = 3.88AQALRR40 pKa = 11.84LRR42 pKa = 11.84LTRR45 pKa = 11.84LNTLYY50 pKa = 11.52NMAGGLNGRR59 pKa = 11.84MQQEE63 pKa = 4.13LRR65 pKa = 11.84DD66 pKa = 3.54LHH68 pKa = 6.82AAIEE72 pKa = 4.16MMDD75 pKa = 4.39LIARR79 pKa = 11.84EE80 pKa = 3.92YY81 pKa = 10.64FINNAEE87 pKa = 3.96ARR89 pKa = 11.84ARR91 pKa = 11.84WAPYY95 pKa = 9.88CAFEE99 pKa = 5.81DD100 pKa = 5.06DD101 pKa = 4.54EE102 pKa = 4.48FHH104 pKa = 7.49HH105 pKa = 7.2LIAEE109 pKa = 4.42ANAGFPSDD117 pKa = 3.82LAIVMEE123 pKa = 4.34PTQEE127 pKa = 4.78DD128 pKa = 3.71EE129 pKa = 4.45DD130 pKa = 4.99DD131 pKa = 4.02RR132 pKa = 11.84DD133 pKa = 3.84SVIDD137 pKa = 3.6ILEE140 pKa = 4.25LTEE143 pKa = 4.68KK144 pKa = 10.54EE145 pKa = 4.17MRR147 pKa = 11.84DD148 pKa = 3.76FMDD151 pKa = 3.13VV152 pKa = 2.91
Molecular weight: 17.61 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5ALV9|A0A0C5ALV9_9VIRU Isolate 1 ORF3 putative replication protein and putative capsid protein genes OS=Phytophthora parasitica virus OX=1608451 PE=4 SV=1
MM1 pKa = 7.38APLVPFHH8 pKa = 6.36TMDD11 pKa = 3.74RR12 pKa = 11.84QWDD15 pKa = 3.43ARR17 pKa = 11.84FNVPTDD23 pKa = 3.54EE24 pKa = 5.04DD25 pKa = 4.55LEE27 pKa = 4.84ALLLNVKK34 pKa = 10.34SEE36 pKa = 3.79VDD38 pKa = 3.09QGKK41 pKa = 8.34FQYY44 pKa = 10.47CLVGGVEE51 pKa = 4.06IGDD54 pKa = 3.45RR55 pKa = 11.84PYY57 pKa = 10.14QTDD60 pKa = 3.57YY61 pKa = 11.11LIKK64 pKa = 9.15HH65 pKa = 4.65VHH67 pKa = 5.3VALVYY72 pKa = 10.54NNRR75 pKa = 11.84VSKK78 pKa = 11.19NSVLKK83 pKa = 10.65NLQIKK88 pKa = 9.0QGNGYY93 pKa = 9.62YY94 pKa = 10.34LVPRR98 pKa = 11.84NRR100 pKa = 11.84DD101 pKa = 3.12LPIQGWIDD109 pKa = 3.12HH110 pKa = 5.78HH111 pKa = 6.51TKK113 pKa = 10.64SKK115 pKa = 10.07TKK117 pKa = 10.16INEE120 pKa = 3.74DD121 pKa = 3.33FKK123 pKa = 11.19LYY125 pKa = 10.62EE126 pKa = 4.41YY127 pKa = 9.21GTLPADD133 pKa = 3.97KK134 pKa = 9.95PNISAQVTKK143 pKa = 10.7RR144 pKa = 11.84SPEE147 pKa = 3.81EE148 pKa = 3.85KK149 pKa = 9.84KK150 pKa = 10.67RR151 pKa = 11.84KK152 pKa = 9.51LDD154 pKa = 3.81DD155 pKa = 4.03VIKK158 pKa = 10.27EE159 pKa = 3.84MRR161 pKa = 11.84GMIEE165 pKa = 3.71EE166 pKa = 4.28GRR168 pKa = 11.84EE169 pKa = 3.65EE170 pKa = 3.78EE171 pKa = 4.77AFTKK175 pKa = 10.43FPRR178 pKa = 11.84NYY180 pKa = 8.62LTYY183 pKa = 10.53GEE185 pKa = 4.83KK186 pKa = 10.15LKK188 pKa = 11.43AMISQRR194 pKa = 11.84RR195 pKa = 11.84DD196 pKa = 3.21FFKK199 pKa = 11.26SNGDD203 pKa = 3.21PHH205 pKa = 7.75IWLTGTPGSGKK216 pKa = 10.08SAILQVIYY224 pKa = 9.42PAYY227 pKa = 10.09YY228 pKa = 10.59NKK230 pKa = 10.47NLDD233 pKa = 3.24NRR235 pKa = 11.84FFDD238 pKa = 4.51LYY240 pKa = 9.83QPKK243 pKa = 9.44VHH245 pKa = 5.45THH247 pKa = 5.74MLLQDD252 pKa = 3.48VDD254 pKa = 3.69HH255 pKa = 6.33TTVEE259 pKa = 4.07KK260 pKa = 11.04LGVQFLKK267 pKa = 9.97TICDD271 pKa = 3.43EE272 pKa = 4.43AGFPVDD278 pKa = 4.12QKK280 pKa = 11.81YY281 pKa = 7.52KK282 pKa = 9.13TPQLARR288 pKa = 11.84TTVLVSSNFTINDD301 pKa = 4.09VIPEE305 pKa = 4.14DD306 pKa = 3.5MPGRR310 pKa = 11.84NEE312 pKa = 3.8NLTALRR318 pKa = 11.84RR319 pKa = 11.84RR320 pKa = 11.84FYY322 pKa = 10.4EE323 pKa = 3.86VNVRR327 pKa = 11.84VLLQILGLKK336 pKa = 9.97LLSKK340 pKa = 11.34YY341 pKa = 9.4EE342 pKa = 4.04LNGLKK347 pKa = 10.14RR348 pKa = 11.84VGNQDD353 pKa = 3.07PTKK356 pKa = 11.06LFMSWDD362 pKa = 3.7YY363 pKa = 11.28TRR365 pKa = 11.84DD366 pKa = 3.31IPTGEE371 pKa = 4.07SLKK374 pKa = 9.76TAKK377 pKa = 10.06EE378 pKa = 3.81YY379 pKa = 10.29QQIIKK384 pKa = 10.16DD385 pKa = 3.71HH386 pKa = 6.4YY387 pKa = 9.56YY388 pKa = 9.14GTNTSPHH395 pKa = 6.31SSQQ398 pKa = 3.59
MM1 pKa = 7.38APLVPFHH8 pKa = 6.36TMDD11 pKa = 3.74RR12 pKa = 11.84QWDD15 pKa = 3.43ARR17 pKa = 11.84FNVPTDD23 pKa = 3.54EE24 pKa = 5.04DD25 pKa = 4.55LEE27 pKa = 4.84ALLLNVKK34 pKa = 10.34SEE36 pKa = 3.79VDD38 pKa = 3.09QGKK41 pKa = 8.34FQYY44 pKa = 10.47CLVGGVEE51 pKa = 4.06IGDD54 pKa = 3.45RR55 pKa = 11.84PYY57 pKa = 10.14QTDD60 pKa = 3.57YY61 pKa = 11.11LIKK64 pKa = 9.15HH65 pKa = 4.65VHH67 pKa = 5.3VALVYY72 pKa = 10.54NNRR75 pKa = 11.84VSKK78 pKa = 11.19NSVLKK83 pKa = 10.65NLQIKK88 pKa = 9.0QGNGYY93 pKa = 9.62YY94 pKa = 10.34LVPRR98 pKa = 11.84NRR100 pKa = 11.84DD101 pKa = 3.12LPIQGWIDD109 pKa = 3.12HH110 pKa = 5.78HH111 pKa = 6.51TKK113 pKa = 10.64SKK115 pKa = 10.07TKK117 pKa = 10.16INEE120 pKa = 3.74DD121 pKa = 3.33FKK123 pKa = 11.19LYY125 pKa = 10.62EE126 pKa = 4.41YY127 pKa = 9.21GTLPADD133 pKa = 3.97KK134 pKa = 9.95PNISAQVTKK143 pKa = 10.7RR144 pKa = 11.84SPEE147 pKa = 3.81EE148 pKa = 3.85KK149 pKa = 9.84KK150 pKa = 10.67RR151 pKa = 11.84KK152 pKa = 9.51LDD154 pKa = 3.81DD155 pKa = 4.03VIKK158 pKa = 10.27EE159 pKa = 3.84MRR161 pKa = 11.84GMIEE165 pKa = 3.71EE166 pKa = 4.28GRR168 pKa = 11.84EE169 pKa = 3.65EE170 pKa = 3.78EE171 pKa = 4.77AFTKK175 pKa = 10.43FPRR178 pKa = 11.84NYY180 pKa = 8.62LTYY183 pKa = 10.53GEE185 pKa = 4.83KK186 pKa = 10.15LKK188 pKa = 11.43AMISQRR194 pKa = 11.84RR195 pKa = 11.84DD196 pKa = 3.21FFKK199 pKa = 11.26SNGDD203 pKa = 3.21PHH205 pKa = 7.75IWLTGTPGSGKK216 pKa = 10.08SAILQVIYY224 pKa = 9.42PAYY227 pKa = 10.09YY228 pKa = 10.59NKK230 pKa = 10.47NLDD233 pKa = 3.24NRR235 pKa = 11.84FFDD238 pKa = 4.51LYY240 pKa = 9.83QPKK243 pKa = 9.44VHH245 pKa = 5.45THH247 pKa = 5.74MLLQDD252 pKa = 3.48VDD254 pKa = 3.69HH255 pKa = 6.33TTVEE259 pKa = 4.07KK260 pKa = 11.04LGVQFLKK267 pKa = 9.97TICDD271 pKa = 3.43EE272 pKa = 4.43AGFPVDD278 pKa = 4.12QKK280 pKa = 11.81YY281 pKa = 7.52KK282 pKa = 9.13TPQLARR288 pKa = 11.84TTVLVSSNFTINDD301 pKa = 4.09VIPEE305 pKa = 4.14DD306 pKa = 3.5MPGRR310 pKa = 11.84NEE312 pKa = 3.8NLTALRR318 pKa = 11.84RR319 pKa = 11.84RR320 pKa = 11.84FYY322 pKa = 10.4EE323 pKa = 3.86VNVRR327 pKa = 11.84VLLQILGLKK336 pKa = 9.97LLSKK340 pKa = 11.34YY341 pKa = 9.4EE342 pKa = 4.04LNGLKK347 pKa = 10.14RR348 pKa = 11.84VGNQDD353 pKa = 3.07PTKK356 pKa = 11.06LFMSWDD362 pKa = 3.7YY363 pKa = 11.28TRR365 pKa = 11.84DD366 pKa = 3.31IPTGEE371 pKa = 4.07SLKK374 pKa = 9.76TAKK377 pKa = 10.06EE378 pKa = 3.81YY379 pKa = 10.29QQIIKK384 pKa = 10.16DD385 pKa = 3.71HH386 pKa = 6.4YY387 pKa = 9.56YY388 pKa = 9.14GTNTSPHH395 pKa = 6.31SSQQ398 pKa = 3.59
Molecular weight: 46.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
996 |
152 |
446 |
332.0 |
37.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.124 ± 1.951 | 0.602 ± 0.05 |
7.229 ± 0.428 | 6.526 ± 1.919 |
3.514 ± 0.233 | 6.627 ± 1.571 |
2.51 ± 0.768 | 4.518 ± 0.625 |
5.823 ± 1.58 | 9.237 ± 0.816 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.108 ± 1.051 | 7.329 ± 1.095 |
4.92 ± 0.947 | 3.916 ± 0.558 |
5.321 ± 0.695 | 4.819 ± 1.03 |
6.124 ± 0.691 | 6.325 ± 0.755 |
1.104 ± 0.177 | 5.321 ± 1.142 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
