
Bosea sp. WAO
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Boseaceae; Bosea; unclassified Bosea
Average proteome isoelectric point is 6.81
Get precalculated fractions of proteins
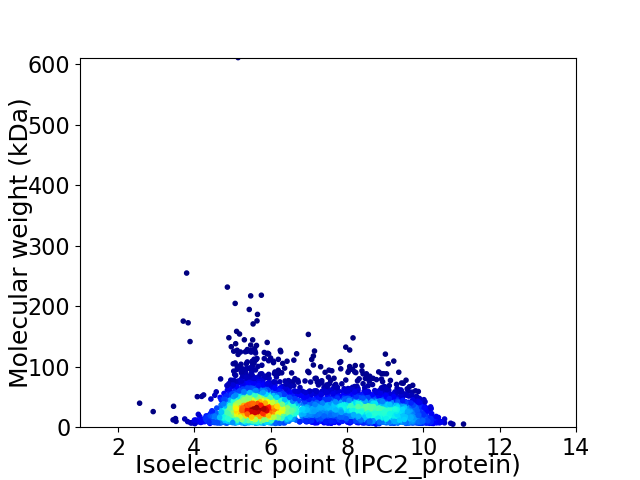
Virtual 2D-PAGE plot for 4978 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
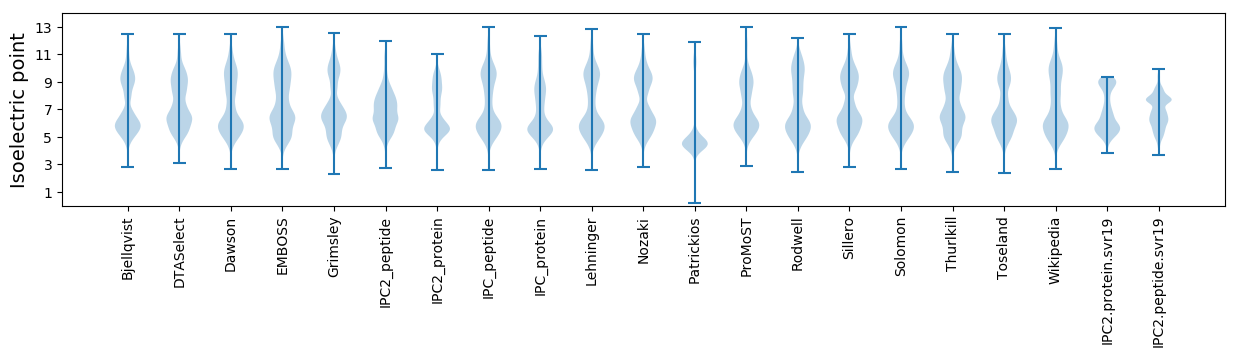
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A124GB96|A0A124GB96_9BRAD Uncharacterized protein OS=Bosea sp. WAO OX=406341 GN=DK26_22450 PE=4 SV=1
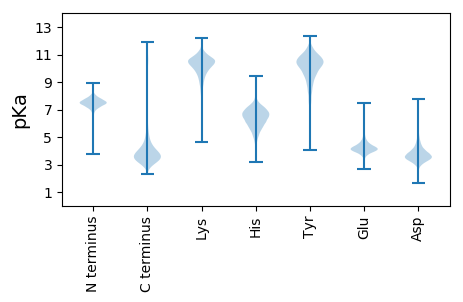
DDD2 pKa = 4.17ATVTEEE8 pKa = 4.47GFGATTGTVGSALSGAYYY26 pKa = 9.39ALTLNADDD34 pKa = 3.56SYYY37 pKa = 10.72YYY39 pKa = 11.59LDDD42 pKa = 3.78SDDD45 pKa = 3.52RR46 pKa = 11.84VNALRR51 pKa = 11.84DDD53 pKa = 3.75EEE55 pKa = 4.44LTEEE59 pKa = 4.04FTYYY63 pKa = 10.59ITDDD67 pKa = 3.4DDD69 pKa = 3.99DDD71 pKa = 4.06EE72 pKa = 4.5TATLTITITGRR83 pKa = 11.84TDDD86 pKa = 2.66TTQIVPGDDD95 pKa = 3.88NGAAHHH101 pKa = 7.04EEE103 pKa = 4.11SVSEEE108 pKa = 4.41GLGDDD113 pKa = 3.95GDDD116 pKa = 4.07SEEE119 pKa = 4.46TTGTVTVTAPDDD131 pKa = 3.63LSTVTVGGRR140 pKa = 11.84VITLAEEE147 pKa = 4.11QALGTSPITVTTPKKK162 pKa = 10.7ILTITGFTPGTTVGGVPIEEE182 pKa = 4.53DDD184 pKa = 3.01SYYY187 pKa = 11.0YYY189 pKa = 11.24EE190 pKa = 3.99TTLQDDD196 pKa = 3.5HH197 pKa = 6.82GGAVDDD203 pKa = 5.98DD204 pKa = 4.33FALEEE209 pKa = 4.32GDDD212 pKa = 4.48GGGSATGTLTIAIADDD228 pKa = 4.22VPQAQDDD235 pKa = 3.25DD236 pKa = 4.2ATIAEEE242 pKa = 4.86DD243 pKa = 3.93DD244 pKa = 4.34SDDD247 pKa = 3.77VDDD250 pKa = 3.23NVFSGVGPGDDD261 pKa = 3.77ADDD264 pKa = 3.94AGNDDD269 pKa = 3.71PATGGPVTGVGFGTTSGTVGSGLSGAYYY297 pKa = 7.67TLTLNADDD305 pKa = 3.47SYYY308 pKa = 10.78YYY310 pKa = 11.54LDDD313 pKa = 3.93GNAAVNGLRR322 pKa = 11.84DDD324 pKa = 3.84QTLTEEE330 pKa = 4.1FSYYY334 pKa = 10.54ISDDD338 pKa = 3.7DDD340 pKa = 3.58STSEEE345 pKa = 4.12TLTITI
DDD2 pKa = 4.17ATVTEEE8 pKa = 4.47GFGATTGTVGSALSGAYYY26 pKa = 9.39ALTLNADDD34 pKa = 3.56SYYY37 pKa = 10.72YYY39 pKa = 11.59LDDD42 pKa = 3.78SDDD45 pKa = 3.52RR46 pKa = 11.84VNALRR51 pKa = 11.84DDD53 pKa = 3.75EEE55 pKa = 4.44LTEEE59 pKa = 4.04FTYYY63 pKa = 10.59ITDDD67 pKa = 3.4DDD69 pKa = 3.99DDD71 pKa = 4.06EE72 pKa = 4.5TATLTITITGRR83 pKa = 11.84TDDD86 pKa = 2.66TTQIVPGDDD95 pKa = 3.88NGAAHHH101 pKa = 7.04EEE103 pKa = 4.11SVSEEE108 pKa = 4.41GLGDDD113 pKa = 3.95GDDD116 pKa = 4.07SEEE119 pKa = 4.46TTGTVTVTAPDDD131 pKa = 3.63LSTVTVGGRR140 pKa = 11.84VITLAEEE147 pKa = 4.11QALGTSPITVTTPKKK162 pKa = 10.7ILTITGFTPGTTVGGVPIEEE182 pKa = 4.53DDD184 pKa = 3.01SYYY187 pKa = 11.0YYY189 pKa = 11.24EE190 pKa = 3.99TTLQDDD196 pKa = 3.5HH197 pKa = 6.82GGAVDDD203 pKa = 5.98DD204 pKa = 4.33FALEEE209 pKa = 4.32GDDD212 pKa = 4.48GGGSATGTLTIAIADDD228 pKa = 4.22VPQAQDDD235 pKa = 3.25DD236 pKa = 4.2ATIAEEE242 pKa = 4.86DD243 pKa = 3.93DD244 pKa = 4.34SDDD247 pKa = 3.77VDDD250 pKa = 3.23NVFSGVGPGDDD261 pKa = 3.77ADDD264 pKa = 3.94AGNDDD269 pKa = 3.71PATGGPVTGVGFGTTSGTVGSGLSGAYYY297 pKa = 7.67TLTLNADDD305 pKa = 3.47SYYY308 pKa = 10.78YYY310 pKa = 11.54LDDD313 pKa = 3.93GNAAVNGLRR322 pKa = 11.84DDD324 pKa = 3.84QTLTEEE330 pKa = 4.1FSYYY334 pKa = 10.54ISDDD338 pKa = 3.7DDD340 pKa = 3.58STSEEE345 pKa = 4.12TLTITI
Molecular weight: 34.57 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A101K3Z8|A0A101K3Z8_9BRAD Gluconolactonase OS=Bosea sp. WAO OX=406341 GN=DK26_03855 PE=4 SV=1
MM1 pKa = 7.43GFSLLKK7 pKa = 10.07TRR9 pKa = 11.84FAQEE13 pKa = 3.34ALGRR17 pKa = 11.84SLAWYY22 pKa = 9.22LRR24 pKa = 11.84LVRR27 pKa = 11.84RR28 pKa = 11.84TNRR31 pKa = 11.84FVVEE35 pKa = 3.74PADD38 pKa = 3.02IYY40 pKa = 11.34EE41 pKa = 4.31RR42 pKa = 11.84VRR44 pKa = 11.84PDD46 pKa = 3.34LPLIIAMWHH55 pKa = 5.87GQHH58 pKa = 6.93IMIPFARR65 pKa = 11.84PDD67 pKa = 3.3WMPACSLVSRR77 pKa = 11.84HH78 pKa = 5.92GDD80 pKa = 3.04GGFNAVALRR89 pKa = 11.84EE90 pKa = 4.09LGIGAIRR97 pKa = 11.84GSGAMGKK104 pKa = 9.51KK105 pKa = 9.61VRR107 pKa = 11.84EE108 pKa = 4.19KK109 pKa = 10.87GGAPAFLAMMRR120 pKa = 11.84RR121 pKa = 11.84LAAGEE126 pKa = 4.24TMVLTADD133 pKa = 3.03IPKK136 pKa = 9.98RR137 pKa = 11.84ARR139 pKa = 11.84VCGAGIVALARR150 pKa = 11.84ASGRR154 pKa = 11.84PIHH157 pKa = 6.79PIAVVTSRR165 pKa = 11.84RR166 pKa = 11.84IDD168 pKa = 3.49FNSWDD173 pKa = 3.8RR174 pKa = 11.84ASIGLPFGRR183 pKa = 11.84GAIVVGEE190 pKa = 4.31AVTVARR196 pKa = 11.84DD197 pKa = 3.26ADD199 pKa = 3.88EE200 pKa = 4.6ATCEE204 pKa = 3.88AARR207 pKa = 11.84LAVQAGLDD215 pKa = 3.67AAHH218 pKa = 6.5EE219 pKa = 3.98RR220 pKa = 11.84AYY222 pKa = 10.92ALIGARR228 pKa = 11.84DD229 pKa = 3.63PGAGLRR235 pKa = 11.84EE236 pKa = 3.63AAAAKK241 pKa = 10.2DD242 pKa = 3.53AATAAAKK249 pKa = 10.17AATPP253 pKa = 3.8
MM1 pKa = 7.43GFSLLKK7 pKa = 10.07TRR9 pKa = 11.84FAQEE13 pKa = 3.34ALGRR17 pKa = 11.84SLAWYY22 pKa = 9.22LRR24 pKa = 11.84LVRR27 pKa = 11.84RR28 pKa = 11.84TNRR31 pKa = 11.84FVVEE35 pKa = 3.74PADD38 pKa = 3.02IYY40 pKa = 11.34EE41 pKa = 4.31RR42 pKa = 11.84VRR44 pKa = 11.84PDD46 pKa = 3.34LPLIIAMWHH55 pKa = 5.87GQHH58 pKa = 6.93IMIPFARR65 pKa = 11.84PDD67 pKa = 3.3WMPACSLVSRR77 pKa = 11.84HH78 pKa = 5.92GDD80 pKa = 3.04GGFNAVALRR89 pKa = 11.84EE90 pKa = 4.09LGIGAIRR97 pKa = 11.84GSGAMGKK104 pKa = 9.51KK105 pKa = 9.61VRR107 pKa = 11.84EE108 pKa = 4.19KK109 pKa = 10.87GGAPAFLAMMRR120 pKa = 11.84RR121 pKa = 11.84LAAGEE126 pKa = 4.24TMVLTADD133 pKa = 3.03IPKK136 pKa = 9.98RR137 pKa = 11.84ARR139 pKa = 11.84VCGAGIVALARR150 pKa = 11.84ASGRR154 pKa = 11.84PIHH157 pKa = 6.79PIAVVTSRR165 pKa = 11.84RR166 pKa = 11.84IDD168 pKa = 3.49FNSWDD173 pKa = 3.8RR174 pKa = 11.84ASIGLPFGRR183 pKa = 11.84GAIVVGEE190 pKa = 4.31AVTVARR196 pKa = 11.84DD197 pKa = 3.26ADD199 pKa = 3.88EE200 pKa = 4.6ATCEE204 pKa = 3.88AARR207 pKa = 11.84LAVQAGLDD215 pKa = 3.67AAHH218 pKa = 6.5EE219 pKa = 3.98RR220 pKa = 11.84AYY222 pKa = 10.92ALIGARR228 pKa = 11.84DD229 pKa = 3.63PGAGLRR235 pKa = 11.84EE236 pKa = 3.63AAAAKK241 pKa = 10.2DD242 pKa = 3.53AATAAAKK249 pKa = 10.17AATPP253 pKa = 3.8
Molecular weight: 26.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1552009 |
41 |
6267 |
311.8 |
33.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.507 ± 0.05 | 0.811 ± 0.012 |
5.174 ± 0.03 | 5.705 ± 0.038 |
3.706 ± 0.026 | 9.017 ± 0.051 |
1.878 ± 0.016 | 5.073 ± 0.024 |
3.227 ± 0.033 | 10.623 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.328 ± 0.018 | 2.358 ± 0.022 |
5.318 ± 0.033 | 3.032 ± 0.017 |
7.212 ± 0.037 | 5.217 ± 0.037 |
5.036 ± 0.039 | 7.406 ± 0.024 |
1.299 ± 0.014 | 2.073 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
