
bacterium 1XD42-54
Taxonomy: cellular organisms; Bacteria; unclassified Bacteria
Average proteome isoelectric point is 6.1
Get precalculated fractions of proteins
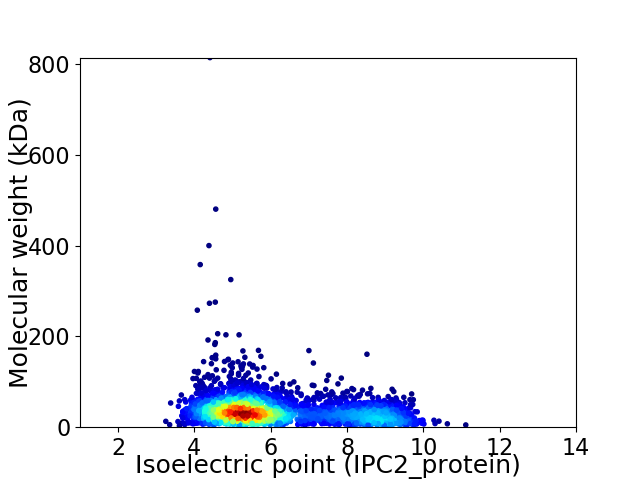
Virtual 2D-PAGE plot for 3209 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
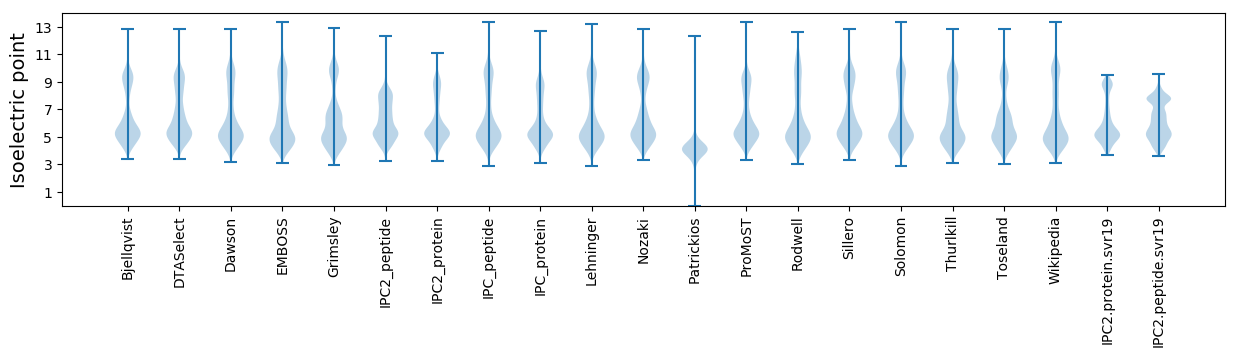
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3A9F9Q8|A0A3A9F9Q8_9BACT GntR family transcriptional regulator OS=bacterium 1XD42-54 OX=2320110 GN=D7Y05_12520 PE=4 SV=1
MM1 pKa = 7.48KK2 pKa = 10.24KK3 pKa = 10.53AIVSVLLATVLLTSGMSVMADD24 pKa = 2.93EE25 pKa = 5.63GDD27 pKa = 3.87TIKK30 pKa = 10.52IGALFNLTGGQASIDD45 pKa = 3.76TPSYY49 pKa = 10.88QGFKK53 pKa = 10.79LKK55 pKa = 10.6ADD57 pKa = 4.81EE58 pKa = 4.32INAAGGINGRR68 pKa = 11.84MIEE71 pKa = 4.19VVSYY75 pKa = 10.8DD76 pKa = 3.4GKK78 pKa = 9.48TDD80 pKa = 3.72QATCANNAKK89 pKa = 10.39KK90 pKa = 10.17MIDD93 pKa = 3.3VDD95 pKa = 3.92DD96 pKa = 4.51CVVVSGFSDD105 pKa = 3.42SNYY108 pKa = 10.7ALAAGAITQDD118 pKa = 3.08KK119 pKa = 9.77QVPFVITGATLPDD132 pKa = 3.65LCEE135 pKa = 3.98QIGDD139 pKa = 3.81YY140 pKa = 11.24SFMVPFGDD148 pKa = 4.22DD149 pKa = 3.0TCAYY153 pKa = 9.94AAAEE157 pKa = 3.96YY158 pKa = 9.97AYY160 pKa = 10.64AEE162 pKa = 4.64IGTKK166 pKa = 9.89CYY168 pKa = 10.66MMIDD172 pKa = 3.28QSMEE176 pKa = 3.88FTKK179 pKa = 10.5TLAEE183 pKa = 4.28CFQEE187 pKa = 4.41RR188 pKa = 11.84FEE190 pKa = 4.23EE191 pKa = 4.32LGGEE195 pKa = 4.17IVGTDD200 pKa = 2.94NYY202 pKa = 10.79MNGDD206 pKa = 4.06PDD208 pKa = 4.82FSAQIDD214 pKa = 3.78KK215 pKa = 10.91FINNDD220 pKa = 2.87GGAEE224 pKa = 4.02FMFFSSVPNDD234 pKa = 3.51AGPLVLQYY242 pKa = 10.39RR243 pKa = 11.84DD244 pKa = 3.74KK245 pKa = 11.28GVEE248 pKa = 3.92LPIVSGDD255 pKa = 3.67GFDD258 pKa = 3.65TPLVAEE264 pKa = 4.32VAGAQSYY271 pKa = 11.13DD272 pKa = 3.58VFVSTHH278 pKa = 5.79CSYY281 pKa = 11.3TNEE284 pKa = 4.12AEE286 pKa = 4.12NVQNFVKK293 pKa = 10.47AYY295 pKa = 7.64TDD297 pKa = 3.34AYY299 pKa = 8.94GTAPEE304 pKa = 4.17NAFAALGYY312 pKa = 10.69DD313 pKa = 3.35AMDD316 pKa = 5.75LIGQALATVEE326 pKa = 5.06DD327 pKa = 4.33PTDD330 pKa = 3.07TAAIKK335 pKa = 10.4DD336 pKa = 4.15AIEE339 pKa = 4.14ATSDD343 pKa = 3.53FKK345 pKa = 11.54GVTGTISYY353 pKa = 9.97SAEE356 pKa = 3.56SHH358 pKa = 6.71KK359 pKa = 10.35PDD361 pKa = 3.08KK362 pKa = 10.03TASILEE368 pKa = 4.21LVDD371 pKa = 5.06GEE373 pKa = 4.49FQFVTEE379 pKa = 4.29EE380 pKa = 3.87
MM1 pKa = 7.48KK2 pKa = 10.24KK3 pKa = 10.53AIVSVLLATVLLTSGMSVMADD24 pKa = 2.93EE25 pKa = 5.63GDD27 pKa = 3.87TIKK30 pKa = 10.52IGALFNLTGGQASIDD45 pKa = 3.76TPSYY49 pKa = 10.88QGFKK53 pKa = 10.79LKK55 pKa = 10.6ADD57 pKa = 4.81EE58 pKa = 4.32INAAGGINGRR68 pKa = 11.84MIEE71 pKa = 4.19VVSYY75 pKa = 10.8DD76 pKa = 3.4GKK78 pKa = 9.48TDD80 pKa = 3.72QATCANNAKK89 pKa = 10.39KK90 pKa = 10.17MIDD93 pKa = 3.3VDD95 pKa = 3.92DD96 pKa = 4.51CVVVSGFSDD105 pKa = 3.42SNYY108 pKa = 10.7ALAAGAITQDD118 pKa = 3.08KK119 pKa = 9.77QVPFVITGATLPDD132 pKa = 3.65LCEE135 pKa = 3.98QIGDD139 pKa = 3.81YY140 pKa = 11.24SFMVPFGDD148 pKa = 4.22DD149 pKa = 3.0TCAYY153 pKa = 9.94AAAEE157 pKa = 3.96YY158 pKa = 9.97AYY160 pKa = 10.64AEE162 pKa = 4.64IGTKK166 pKa = 9.89CYY168 pKa = 10.66MMIDD172 pKa = 3.28QSMEE176 pKa = 3.88FTKK179 pKa = 10.5TLAEE183 pKa = 4.28CFQEE187 pKa = 4.41RR188 pKa = 11.84FEE190 pKa = 4.23EE191 pKa = 4.32LGGEE195 pKa = 4.17IVGTDD200 pKa = 2.94NYY202 pKa = 10.79MNGDD206 pKa = 4.06PDD208 pKa = 4.82FSAQIDD214 pKa = 3.78KK215 pKa = 10.91FINNDD220 pKa = 2.87GGAEE224 pKa = 4.02FMFFSSVPNDD234 pKa = 3.51AGPLVLQYY242 pKa = 10.39RR243 pKa = 11.84DD244 pKa = 3.74KK245 pKa = 11.28GVEE248 pKa = 3.92LPIVSGDD255 pKa = 3.67GFDD258 pKa = 3.65TPLVAEE264 pKa = 4.32VAGAQSYY271 pKa = 11.13DD272 pKa = 3.58VFVSTHH278 pKa = 5.79CSYY281 pKa = 11.3TNEE284 pKa = 4.12AEE286 pKa = 4.12NVQNFVKK293 pKa = 10.47AYY295 pKa = 7.64TDD297 pKa = 3.34AYY299 pKa = 8.94GTAPEE304 pKa = 4.17NAFAALGYY312 pKa = 10.69DD313 pKa = 3.35AMDD316 pKa = 5.75LIGQALATVEE326 pKa = 5.06DD327 pKa = 4.33PTDD330 pKa = 3.07TAAIKK335 pKa = 10.4DD336 pKa = 4.15AIEE339 pKa = 4.14ATSDD343 pKa = 3.53FKK345 pKa = 11.54GVTGTISYY353 pKa = 9.97SAEE356 pKa = 3.56SHH358 pKa = 6.71KK359 pKa = 10.35PDD361 pKa = 3.08KK362 pKa = 10.03TASILEE368 pKa = 4.21LVDD371 pKa = 5.06GEE373 pKa = 4.49FQFVTEE379 pKa = 4.29EE380 pKa = 3.87
Molecular weight: 40.55 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3A9FCM9|A0A3A9FCM9_9BACT Dephospho-CoA kinase OS=bacterium 1XD42-54 OX=2320110 GN=coaE PE=3 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 9.06KK9 pKa = 8.03RR10 pKa = 11.84SRR12 pKa = 11.84SKK14 pKa = 9.44VHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTAGGRR28 pKa = 11.84KK29 pKa = 8.71VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.71GRR39 pKa = 11.84KK40 pKa = 8.73VLSAA44 pKa = 4.05
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 9.06KK9 pKa = 8.03RR10 pKa = 11.84SRR12 pKa = 11.84SKK14 pKa = 9.44VHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTAGGRR28 pKa = 11.84KK29 pKa = 8.71VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.71GRR39 pKa = 11.84KK40 pKa = 8.73VLSAA44 pKa = 4.05
Molecular weight: 4.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1081669 |
17 |
7285 |
337.1 |
37.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.824 ± 0.043 | 1.533 ± 0.021 |
5.406 ± 0.038 | 7.896 ± 0.051 |
4.046 ± 0.034 | 7.304 ± 0.05 |
1.719 ± 0.016 | 6.821 ± 0.039 |
6.176 ± 0.043 | 9.167 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.065 ± 0.026 | 4.009 ± 0.029 |
3.274 ± 0.026 | 3.619 ± 0.029 |
4.816 ± 0.041 | 5.769 ± 0.035 |
5.489 ± 0.055 | 6.951 ± 0.035 |
0.957 ± 0.012 | 4.16 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
