
spirochete symbiont of Stewartia floridana
Taxonomy: cellular organisms; Bacteria; Spirochaetes; Spirochaetia; Spirochaetales; unclassified Spirochaetales
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
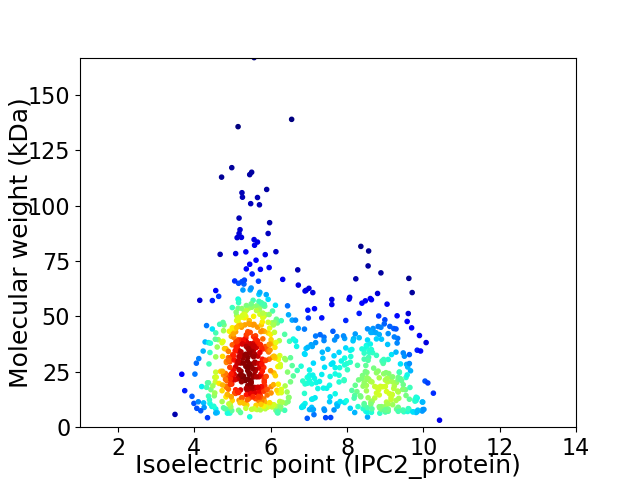
Virtual 2D-PAGE plot for 817 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A421K8W2|A0A421K8W2_9SPIR Uncharacterized protein OS=spirochete symbiont of Stewartia floridana OX=1968597 GN=B6D68_03135 PE=4 SV=1
MM1 pKa = 7.66KK2 pKa = 10.15NYY4 pKa = 10.45QNITLSLLVFTLFWGCQASLPDD26 pKa = 3.6NLEE29 pKa = 4.01TLGEE33 pKa = 4.35SGLAWEE39 pKa = 4.78MSTLTSEE46 pKa = 5.38DD47 pKa = 3.81DD48 pKa = 4.39LNVLLARR55 pKa = 11.84SGEE58 pKa = 4.41GIVDD62 pKa = 3.45WQLSRR67 pKa = 11.84EE68 pKa = 4.14FAAMALEE75 pKa = 4.03EE76 pKa = 5.25FIEE79 pKa = 4.3EE80 pKa = 4.05DD81 pKa = 3.38NYY83 pKa = 10.98PPEE86 pKa = 5.16SEE88 pKa = 4.77LWDD91 pKa = 3.61IPIAIYY97 pKa = 10.56DD98 pKa = 3.77DD99 pKa = 3.91EE100 pKa = 6.46GEE102 pKa = 3.95VRR104 pKa = 11.84YY105 pKa = 10.27YY106 pKa = 10.42EE107 pKa = 3.95FRR109 pKa = 11.84IVDD112 pKa = 3.92GEE114 pKa = 4.24QTLGAIAGNARR125 pKa = 11.84EE126 pKa = 4.33DD127 pKa = 3.13RR128 pKa = 11.84GAPVSHH134 pKa = 6.2VFEE137 pKa = 4.18MDD139 pKa = 3.57GYY141 pKa = 11.19ADD143 pKa = 4.12EE144 pKa = 5.72LSALYY149 pKa = 10.58ASGAMSADD157 pKa = 4.43DD158 pKa = 3.72IPRR161 pKa = 11.84IVDD164 pKa = 3.24NDD166 pKa = 3.71YY167 pKa = 10.79PSYY170 pKa = 11.26AVGSVTLTRR179 pKa = 11.84SGNVDD184 pKa = 3.3LKK186 pKa = 11.11EE187 pKa = 4.06IVDD190 pKa = 4.0PSTGEE195 pKa = 3.65QLEE198 pKa = 4.29NLVVLLDD205 pKa = 3.83MEE207 pKa = 4.49EE208 pKa = 4.66SIAAYY213 pKa = 9.57PDD215 pKa = 3.41TT216 pKa = 4.74
MM1 pKa = 7.66KK2 pKa = 10.15NYY4 pKa = 10.45QNITLSLLVFTLFWGCQASLPDD26 pKa = 3.6NLEE29 pKa = 4.01TLGEE33 pKa = 4.35SGLAWEE39 pKa = 4.78MSTLTSEE46 pKa = 5.38DD47 pKa = 3.81DD48 pKa = 4.39LNVLLARR55 pKa = 11.84SGEE58 pKa = 4.41GIVDD62 pKa = 3.45WQLSRR67 pKa = 11.84EE68 pKa = 4.14FAAMALEE75 pKa = 4.03EE76 pKa = 5.25FIEE79 pKa = 4.3EE80 pKa = 4.05DD81 pKa = 3.38NYY83 pKa = 10.98PPEE86 pKa = 5.16SEE88 pKa = 4.77LWDD91 pKa = 3.61IPIAIYY97 pKa = 10.56DD98 pKa = 3.77DD99 pKa = 3.91EE100 pKa = 6.46GEE102 pKa = 3.95VRR104 pKa = 11.84YY105 pKa = 10.27YY106 pKa = 10.42EE107 pKa = 3.95FRR109 pKa = 11.84IVDD112 pKa = 3.92GEE114 pKa = 4.24QTLGAIAGNARR125 pKa = 11.84EE126 pKa = 4.33DD127 pKa = 3.13RR128 pKa = 11.84GAPVSHH134 pKa = 6.2VFEE137 pKa = 4.18MDD139 pKa = 3.57GYY141 pKa = 11.19ADD143 pKa = 4.12EE144 pKa = 5.72LSALYY149 pKa = 10.58ASGAMSADD157 pKa = 4.43DD158 pKa = 3.72IPRR161 pKa = 11.84IVDD164 pKa = 3.24NDD166 pKa = 3.71YY167 pKa = 10.79PSYY170 pKa = 11.26AVGSVTLTRR179 pKa = 11.84SGNVDD184 pKa = 3.3LKK186 pKa = 11.11EE187 pKa = 4.06IVDD190 pKa = 4.0PSTGEE195 pKa = 3.65QLEE198 pKa = 4.29NLVVLLDD205 pKa = 3.83MEE207 pKa = 4.49EE208 pKa = 4.66SIAAYY213 pKa = 9.57PDD215 pKa = 3.41TT216 pKa = 4.74
Molecular weight: 23.91 kDa
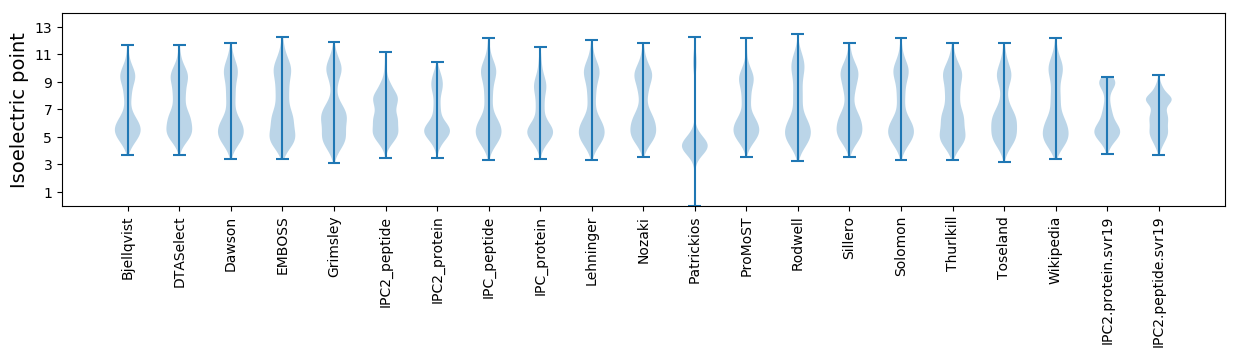
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A421K9E7|A0A421K9E7_9SPIR Uncharacterized protein OS=spirochete symbiont of Stewartia floridana OX=1968597 GN=B6D68_02115 PE=4 SV=1
MM1 pKa = 7.08SQTRR5 pKa = 11.84SLAGLLISIVFMAILCSPQALHH27 pKa = 7.01PVLQTLFPQAGEE39 pKa = 4.13WVFPRR44 pKa = 11.84ATLIRR49 pKa = 11.84LTIQHH54 pKa = 6.48LTLSLTAAGIAALIGILLAITATRR78 pKa = 11.84RR79 pKa = 11.84HH80 pKa = 5.12GRR82 pKa = 11.84AFLRR86 pKa = 11.84MIRR89 pKa = 11.84DD90 pKa = 3.69VSSLAQTIPPTAVLALAIPFLGFGYY115 pKa = 10.4KK116 pKa = 9.64PVLLALVLYY125 pKa = 10.67SILPVLNNTLTALEE139 pKa = 4.81CLPKK143 pKa = 10.31PILEE147 pKa = 4.56AAQAMGMKK155 pKa = 9.88SLQVLFLIEE164 pKa = 4.8LPLALPAIGTGIRR177 pKa = 11.84ISAVINIGTATIAALAGAGGLGSPIVAGLVRR208 pKa = 11.84DD209 pKa = 3.76NPAWVFQGAASSALLALILNAILLRR234 pKa = 11.84VEE236 pKa = 4.78VKK238 pKa = 10.79LNPAKK243 pKa = 10.71
MM1 pKa = 7.08SQTRR5 pKa = 11.84SLAGLLISIVFMAILCSPQALHH27 pKa = 7.01PVLQTLFPQAGEE39 pKa = 4.13WVFPRR44 pKa = 11.84ATLIRR49 pKa = 11.84LTIQHH54 pKa = 6.48LTLSLTAAGIAALIGILLAITATRR78 pKa = 11.84RR79 pKa = 11.84HH80 pKa = 5.12GRR82 pKa = 11.84AFLRR86 pKa = 11.84MIRR89 pKa = 11.84DD90 pKa = 3.69VSSLAQTIPPTAVLALAIPFLGFGYY115 pKa = 10.4KK116 pKa = 9.64PVLLALVLYY125 pKa = 10.67SILPVLNNTLTALEE139 pKa = 4.81CLPKK143 pKa = 10.31PILEE147 pKa = 4.56AAQAMGMKK155 pKa = 9.88SLQVLFLIEE164 pKa = 4.8LPLALPAIGTGIRR177 pKa = 11.84ISAVINIGTATIAALAGAGGLGSPIVAGLVRR208 pKa = 11.84DD209 pKa = 3.76NPAWVFQGAASSALLALILNAILLRR234 pKa = 11.84VEE236 pKa = 4.78VKK238 pKa = 10.79LNPAKK243 pKa = 10.71
Molecular weight: 25.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
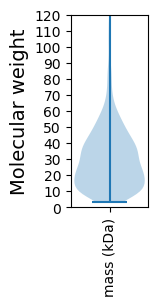
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
230933 |
27 |
1508 |
282.7 |
31.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.175 ± 0.093 | 1.121 ± 0.037 |
5.492 ± 0.065 | 6.437 ± 0.068 |
3.978 ± 0.06 | 8.227 ± 0.068 |
2.131 ± 0.039 | 6.909 ± 0.067 |
4.527 ± 0.067 | 10.049 ± 0.097 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.724 ± 0.041 | 3.432 ± 0.046 |
4.64 ± 0.058 | 2.901 ± 0.046 |
6.537 ± 0.075 | 6.479 ± 0.055 |
4.694 ± 0.054 | 6.462 ± 0.071 |
1.287 ± 0.034 | 2.796 ± 0.051 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
