
Lactiplantibacillus mudanjiangensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Lactiplantibacillus
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
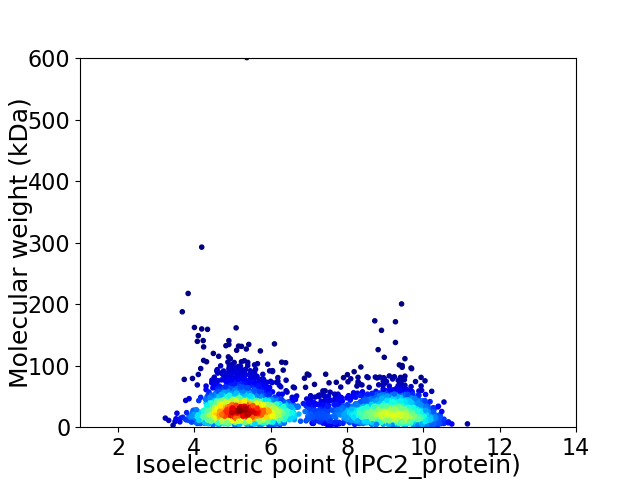
Virtual 2D-PAGE plot for 3503 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A660E4B9|A0A660E4B9_9LACO DNA-binding protein XRE family [Lactobacillus sakei subsp. sakei 23K] OS=Lactiplantibacillus mudanjiangensis OX=1296538 GN=MUDAN_MDHGFNIF_01789 PE=4 SV=1
MM1 pKa = 7.95ADD3 pKa = 3.39QLVLEE8 pKa = 4.39TQKK11 pKa = 10.47WLNATYY17 pKa = 11.07GDD19 pKa = 3.61VSGFGSVPEE28 pKa = 4.83DD29 pKa = 4.47GFTGWDD35 pKa = 3.12TVYY38 pKa = 11.3GLTRR42 pKa = 11.84ALQHH46 pKa = 6.13EE47 pKa = 4.73LGITDD52 pKa = 4.26LVDD55 pKa = 3.26NFGPTTASLFDD66 pKa = 4.77EE67 pKa = 4.75IADD70 pKa = 4.29EE71 pKa = 4.2IVPGYY76 pKa = 9.57SGNIAYY82 pKa = 9.53IIQGGFWCKK91 pKa = 10.65GIDD94 pKa = 3.4PVAFDD99 pKa = 5.08GEE101 pKa = 4.5FSSDD105 pKa = 3.25TEE107 pKa = 4.56SAVSTMKK114 pKa = 10.47EE115 pKa = 3.82YY116 pKa = 10.99AGLSDD121 pKa = 3.77TSGTLNSQFMAALLNMSAFTLVADD145 pKa = 5.1GYY147 pKa = 10.3EE148 pKa = 4.68KK149 pKa = 10.39IRR151 pKa = 11.84TMQQQLNHH159 pKa = 7.5DD160 pKa = 4.0YY161 pKa = 10.93LAYY164 pKa = 9.73TGILPCDD171 pKa = 4.69GIYY174 pKa = 10.55QRR176 pKa = 11.84DD177 pKa = 3.81TNTALLYY184 pKa = 10.65ALQAEE189 pKa = 4.68EE190 pKa = 5.61GLDD193 pKa = 3.43TDD195 pKa = 4.49TASGTYY201 pKa = 10.85GPMTQEE207 pKa = 3.89LTPTVYY213 pKa = 10.71EE214 pKa = 4.19GDD216 pKa = 3.56TNNFVRR222 pKa = 11.84ILQWGLYY229 pKa = 9.56VNNQTYY235 pKa = 9.41TGDD238 pKa = 3.41FDD240 pKa = 3.77GTYY243 pKa = 10.86SLDD246 pKa = 3.32VSSAVSTFEE255 pKa = 3.84TDD257 pKa = 2.81MALPSTSGASAGLDD271 pKa = 3.05VFMSLLTSAGNPNRR285 pKa = 11.84AAVACDD291 pKa = 3.21TSHH294 pKa = 7.23QINASEE300 pKa = 3.94AAALNTAGYY309 pKa = 9.23EE310 pKa = 4.01YY311 pKa = 10.8VGRR314 pKa = 11.84YY315 pKa = 7.02LTGTAGTGDD324 pKa = 3.56NEE326 pKa = 4.24VAKK329 pKa = 10.77NLTTTEE335 pKa = 3.77ISTLTDD341 pKa = 3.06AGLKK345 pKa = 9.86IFPIYY350 pKa = 9.99QDD352 pKa = 3.47GASDD356 pKa = 3.79SEE358 pKa = 5.21DD359 pKa = 3.67YY360 pKa = 9.54FTEE363 pKa = 4.38AKK365 pKa = 10.3GKK367 pKa = 10.27SDD369 pKa = 3.17GRR371 pKa = 11.84KK372 pKa = 9.43AGLAAMEE379 pKa = 5.15LGFPDD384 pKa = 4.31DD385 pKa = 4.63AIIYY389 pKa = 7.6FAVDD393 pKa = 2.75VDD395 pKa = 3.88IQDD398 pKa = 3.32GDD400 pKa = 3.54IAGTIAPYY408 pKa = 8.63FTGVASGIAGFDD420 pKa = 3.81FNVGIYY426 pKa = 8.04ATRR429 pKa = 11.84NVVNTIITDD438 pKa = 4.14GLAEE442 pKa = 4.26KK443 pKa = 10.83AFVSDD448 pKa = 3.63MSTGFSGNLGFAMPTEE464 pKa = 3.94WAFDD468 pKa = 3.57QFSEE472 pKa = 4.72VAFDD476 pKa = 4.93DD477 pKa = 4.82FYY479 pKa = 11.11IDD481 pKa = 3.9KK482 pKa = 10.61VASNSSRR489 pKa = 11.84STAVSDD495 pKa = 4.08FTAGGAVGGADD506 pKa = 4.2DD507 pKa = 4.12LAKK510 pKa = 10.41INEE513 pKa = 4.57IISDD517 pKa = 4.05LGDD520 pKa = 3.32TDD522 pKa = 5.66SIFNFVKK529 pKa = 10.35DD530 pKa = 3.78VKK532 pKa = 10.7IEE534 pKa = 4.06ATSEE538 pKa = 4.26EE539 pKa = 4.77YY540 pKa = 10.64KK541 pKa = 9.79VTTPAMDD548 pKa = 4.53LSLKK552 pKa = 10.32VDD554 pKa = 4.13LEE556 pKa = 4.38GSISDD561 pKa = 4.42DD562 pKa = 4.74DD563 pKa = 5.25GDD565 pKa = 3.78TSVTYY570 pKa = 10.42NVNDD574 pKa = 4.08GSVHH578 pKa = 6.48VDD580 pKa = 3.16LGDD583 pKa = 5.78DD584 pKa = 3.55ISQLLAEE591 pKa = 4.41NTAGFTDD598 pKa = 3.85TAVTASLDD606 pKa = 3.81SVGSVIKK613 pKa = 10.86NGEE616 pKa = 4.07IKK618 pKa = 10.98NKK620 pKa = 9.73IDD622 pKa = 3.36VSATATTITFKK633 pKa = 10.91LDD635 pKa = 3.1STFEE639 pKa = 4.15HH640 pKa = 7.05EE641 pKa = 4.74SNNGKK646 pKa = 8.07TYY648 pKa = 9.92EE649 pKa = 3.94IEE651 pKa = 4.11YY652 pKa = 10.32EE653 pKa = 3.82ITLVVSFHH661 pKa = 7.11SIITPLPDD669 pKa = 3.86WITATEE675 pKa = 3.86ADD677 pKa = 3.66AHH679 pKa = 6.1NSALTEE685 pKa = 3.96IQGDD689 pKa = 3.64AVRR692 pKa = 11.84IMEE695 pKa = 4.5GAAVVVGLAIGAIAIAGLIASGTFVVGYY723 pKa = 10.15LVDD726 pKa = 4.11AVALAVGAII735 pKa = 3.8
MM1 pKa = 7.95ADD3 pKa = 3.39QLVLEE8 pKa = 4.39TQKK11 pKa = 10.47WLNATYY17 pKa = 11.07GDD19 pKa = 3.61VSGFGSVPEE28 pKa = 4.83DD29 pKa = 4.47GFTGWDD35 pKa = 3.12TVYY38 pKa = 11.3GLTRR42 pKa = 11.84ALQHH46 pKa = 6.13EE47 pKa = 4.73LGITDD52 pKa = 4.26LVDD55 pKa = 3.26NFGPTTASLFDD66 pKa = 4.77EE67 pKa = 4.75IADD70 pKa = 4.29EE71 pKa = 4.2IVPGYY76 pKa = 9.57SGNIAYY82 pKa = 9.53IIQGGFWCKK91 pKa = 10.65GIDD94 pKa = 3.4PVAFDD99 pKa = 5.08GEE101 pKa = 4.5FSSDD105 pKa = 3.25TEE107 pKa = 4.56SAVSTMKK114 pKa = 10.47EE115 pKa = 3.82YY116 pKa = 10.99AGLSDD121 pKa = 3.77TSGTLNSQFMAALLNMSAFTLVADD145 pKa = 5.1GYY147 pKa = 10.3EE148 pKa = 4.68KK149 pKa = 10.39IRR151 pKa = 11.84TMQQQLNHH159 pKa = 7.5DD160 pKa = 4.0YY161 pKa = 10.93LAYY164 pKa = 9.73TGILPCDD171 pKa = 4.69GIYY174 pKa = 10.55QRR176 pKa = 11.84DD177 pKa = 3.81TNTALLYY184 pKa = 10.65ALQAEE189 pKa = 4.68EE190 pKa = 5.61GLDD193 pKa = 3.43TDD195 pKa = 4.49TASGTYY201 pKa = 10.85GPMTQEE207 pKa = 3.89LTPTVYY213 pKa = 10.71EE214 pKa = 4.19GDD216 pKa = 3.56TNNFVRR222 pKa = 11.84ILQWGLYY229 pKa = 9.56VNNQTYY235 pKa = 9.41TGDD238 pKa = 3.41FDD240 pKa = 3.77GTYY243 pKa = 10.86SLDD246 pKa = 3.32VSSAVSTFEE255 pKa = 3.84TDD257 pKa = 2.81MALPSTSGASAGLDD271 pKa = 3.05VFMSLLTSAGNPNRR285 pKa = 11.84AAVACDD291 pKa = 3.21TSHH294 pKa = 7.23QINASEE300 pKa = 3.94AAALNTAGYY309 pKa = 9.23EE310 pKa = 4.01YY311 pKa = 10.8VGRR314 pKa = 11.84YY315 pKa = 7.02LTGTAGTGDD324 pKa = 3.56NEE326 pKa = 4.24VAKK329 pKa = 10.77NLTTTEE335 pKa = 3.77ISTLTDD341 pKa = 3.06AGLKK345 pKa = 9.86IFPIYY350 pKa = 9.99QDD352 pKa = 3.47GASDD356 pKa = 3.79SEE358 pKa = 5.21DD359 pKa = 3.67YY360 pKa = 9.54FTEE363 pKa = 4.38AKK365 pKa = 10.3GKK367 pKa = 10.27SDD369 pKa = 3.17GRR371 pKa = 11.84KK372 pKa = 9.43AGLAAMEE379 pKa = 5.15LGFPDD384 pKa = 4.31DD385 pKa = 4.63AIIYY389 pKa = 7.6FAVDD393 pKa = 2.75VDD395 pKa = 3.88IQDD398 pKa = 3.32GDD400 pKa = 3.54IAGTIAPYY408 pKa = 8.63FTGVASGIAGFDD420 pKa = 3.81FNVGIYY426 pKa = 8.04ATRR429 pKa = 11.84NVVNTIITDD438 pKa = 4.14GLAEE442 pKa = 4.26KK443 pKa = 10.83AFVSDD448 pKa = 3.63MSTGFSGNLGFAMPTEE464 pKa = 3.94WAFDD468 pKa = 3.57QFSEE472 pKa = 4.72VAFDD476 pKa = 4.93DD477 pKa = 4.82FYY479 pKa = 11.11IDD481 pKa = 3.9KK482 pKa = 10.61VASNSSRR489 pKa = 11.84STAVSDD495 pKa = 4.08FTAGGAVGGADD506 pKa = 4.2DD507 pKa = 4.12LAKK510 pKa = 10.41INEE513 pKa = 4.57IISDD517 pKa = 4.05LGDD520 pKa = 3.32TDD522 pKa = 5.66SIFNFVKK529 pKa = 10.35DD530 pKa = 3.78VKK532 pKa = 10.7IEE534 pKa = 4.06ATSEE538 pKa = 4.26EE539 pKa = 4.77YY540 pKa = 10.64KK541 pKa = 9.79VTTPAMDD548 pKa = 4.53LSLKK552 pKa = 10.32VDD554 pKa = 4.13LEE556 pKa = 4.38GSISDD561 pKa = 4.42DD562 pKa = 4.74DD563 pKa = 5.25GDD565 pKa = 3.78TSVTYY570 pKa = 10.42NVNDD574 pKa = 4.08GSVHH578 pKa = 6.48VDD580 pKa = 3.16LGDD583 pKa = 5.78DD584 pKa = 3.55ISQLLAEE591 pKa = 4.41NTAGFTDD598 pKa = 3.85TAVTASLDD606 pKa = 3.81SVGSVIKK613 pKa = 10.86NGEE616 pKa = 4.07IKK618 pKa = 10.98NKK620 pKa = 9.73IDD622 pKa = 3.36VSATATTITFKK633 pKa = 10.91LDD635 pKa = 3.1STFEE639 pKa = 4.15HH640 pKa = 7.05EE641 pKa = 4.74SNNGKK646 pKa = 8.07TYY648 pKa = 9.92EE649 pKa = 3.94IEE651 pKa = 4.11YY652 pKa = 10.32EE653 pKa = 3.82ITLVVSFHH661 pKa = 7.11SIITPLPDD669 pKa = 3.86WITATEE675 pKa = 3.86ADD677 pKa = 3.66AHH679 pKa = 6.1NSALTEE685 pKa = 3.96IQGDD689 pKa = 3.64AVRR692 pKa = 11.84IMEE695 pKa = 4.5GAAVVVGLAIGAIAIAGLIASGTFVVGYY723 pKa = 10.15LVDD726 pKa = 4.11AVALAVGAII735 pKa = 3.8
Molecular weight: 77.71 kDa
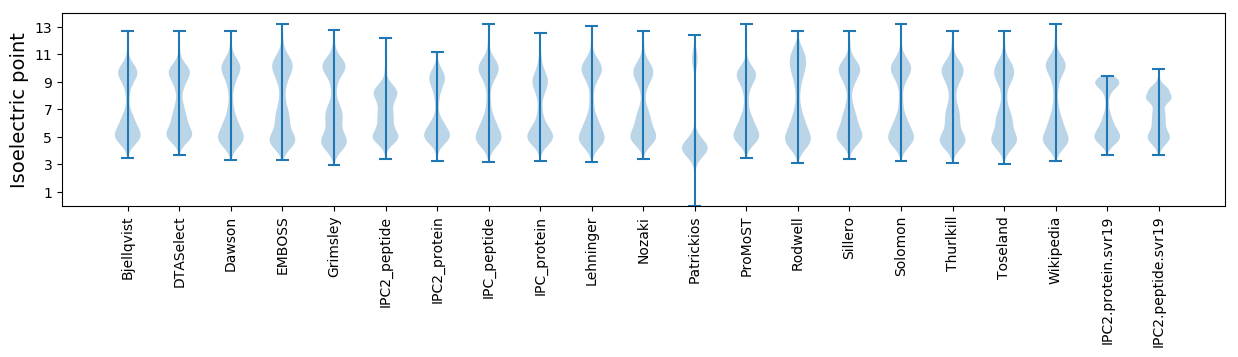
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A660DY40|A0A660DY40_9LACO Uncharacterized protein OS=Lactiplantibacillus mudanjiangensis OX=1296538 GN=MUDAN_MDHGFNIF_00232 PE=4 SV=1
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.69QPKK8 pKa = 8.69KK9 pKa = 7.77RR10 pKa = 11.84HH11 pKa = 4.99RR12 pKa = 11.84QRR14 pKa = 11.84VHH16 pKa = 6.07GFRR19 pKa = 11.84KK20 pKa = 10.03RR21 pKa = 11.84MSTSNGRR28 pKa = 11.84KK29 pKa = 8.01VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84QRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.8VLSAA44 pKa = 4.11
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.69QPKK8 pKa = 8.69KK9 pKa = 7.77RR10 pKa = 11.84HH11 pKa = 4.99RR12 pKa = 11.84QRR14 pKa = 11.84VHH16 pKa = 6.07GFRR19 pKa = 11.84KK20 pKa = 10.03RR21 pKa = 11.84MSTSNGRR28 pKa = 11.84KK29 pKa = 8.01VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84QRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.8VLSAA44 pKa = 4.11
Molecular weight: 5.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1003855 |
29 |
5304 |
286.6 |
31.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.108 ± 0.052 | 0.466 ± 0.01 |
5.684 ± 0.05 | 4.423 ± 0.047 |
3.972 ± 0.029 | 6.643 ± 0.04 |
2.254 ± 0.022 | 6.362 ± 0.038 |
5.569 ± 0.05 | 10.159 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.656 ± 0.024 | 4.383 ± 0.033 |
3.729 ± 0.023 | 5.324 ± 0.045 |
3.99 ± 0.037 | 5.767 ± 0.048 |
7.438 ± 0.074 | 7.27 ± 0.033 |
1.239 ± 0.02 | 3.565 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
