
Nocardioides anomalus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Propionibacteriales; Nocardioidaceae; Nocardioides
Average proteome isoelectric point is 5.93
Get precalculated fractions of proteins
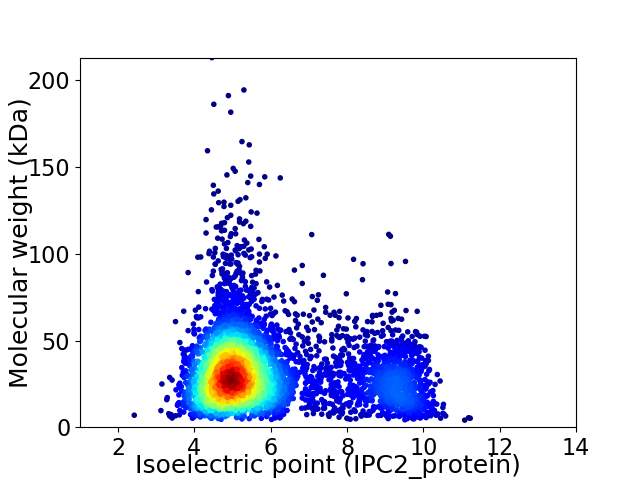
Virtual 2D-PAGE plot for 4914 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
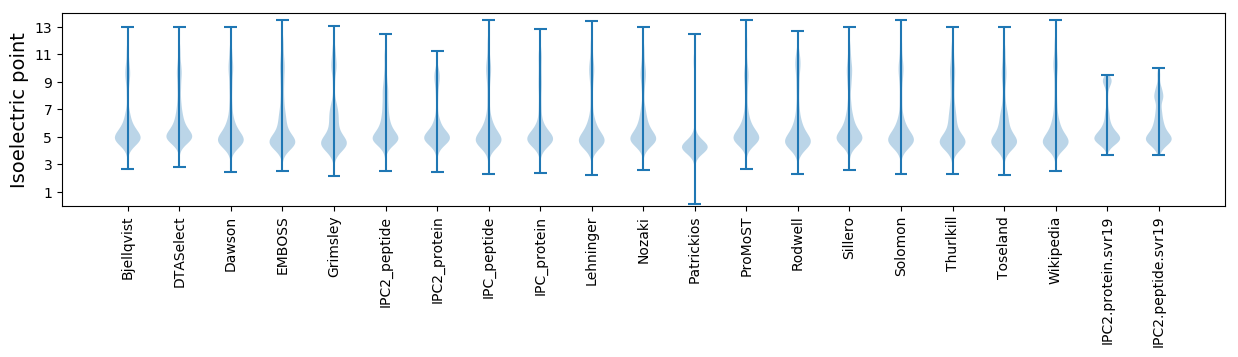
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6G6WKQ3|A0A6G6WKQ3_9ACTN DNA internalization-related competence protein ComEC/Rec2 OS=Nocardioides anomalus OX=2712223 GN=G5V58_07830 PE=4 SV=1
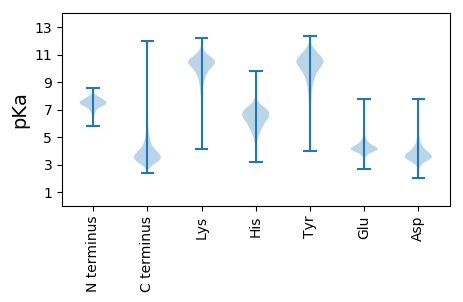
MM1 pKa = 7.16GAALGFSLVACGDD14 pKa = 4.02DD15 pKa = 4.23SDD17 pKa = 5.88SGDD20 pKa = 3.69DD21 pKa = 3.54TSGSDD26 pKa = 3.51EE27 pKa = 4.38SSSASADD34 pKa = 3.46TSEE37 pKa = 4.59SASEE41 pKa = 4.2SASADD46 pKa = 3.13AGGSFTNDD54 pKa = 2.31KK55 pKa = 10.72CAGSDD60 pKa = 3.08TSANKK65 pKa = 9.76FAVGGILPLTGNLAFLGPPAVAGVGLAVSDD95 pKa = 3.7INAAGGVNGAQACSNILDD113 pKa = 4.54SGDD116 pKa = 3.73STDD119 pKa = 3.8MSISTASAGTMVAAKK134 pKa = 9.2PSVVIGAEE142 pKa = 4.09SSSVSLNFVDD152 pKa = 4.02TLTDD156 pKa = 3.42AKK158 pKa = 9.61ITQISPANTAIDD170 pKa = 3.83LSGYY174 pKa = 9.48SPFYY178 pKa = 10.71FRR180 pKa = 11.84TAPPDD185 pKa = 3.84TVQGNALGTLISQDD199 pKa = 3.15GYY201 pKa = 9.09TNIGFLVFNDD211 pKa = 3.95TYY213 pKa = 11.22GTGLRR218 pKa = 11.84NAVQQTVEE226 pKa = 3.91AAGGKK231 pKa = 9.05CVYY234 pKa = 9.91GCKK237 pKa = 10.13GDD239 pKa = 4.08GDD241 pKa = 4.34EE242 pKa = 5.39FPAGQTTFSAEE253 pKa = 4.16VQAVTAAKK261 pKa = 9.51PDD263 pKa = 4.43AIVIIAFDD271 pKa = 3.8EE272 pKa = 4.54TKK274 pKa = 10.81SIVPEE279 pKa = 4.08LASSGFDD286 pKa = 3.01MSKK289 pKa = 9.61TYY291 pKa = 10.4FVDD294 pKa = 4.82GNLSDD299 pKa = 3.94YY300 pKa = 11.2GQDD303 pKa = 3.73FEE305 pKa = 5.7PGTLEE310 pKa = 4.3GAQGTLPGQDD320 pKa = 3.56PDD322 pKa = 4.72QGFKK326 pKa = 11.28DD327 pKa = 4.64NLTGWYY333 pKa = 9.79KK334 pKa = 10.48SANGEE339 pKa = 4.1DD340 pKa = 3.81LKK342 pKa = 11.15DD343 pKa = 3.54YY344 pKa = 11.1SYY346 pKa = 11.63GAEE349 pKa = 4.32SYY351 pKa = 10.87DD352 pKa = 3.59ATILAALAAAKK363 pKa = 10.41GGANDD368 pKa = 3.56SQTIQKK374 pKa = 9.87NLAAVSGATDD384 pKa = 3.78GEE386 pKa = 4.49EE387 pKa = 3.89CSTYY391 pKa = 11.22ADD393 pKa = 3.8CLKK396 pKa = 10.63LIEE399 pKa = 4.98GGSDD403 pKa = 2.54IHH405 pKa = 5.63YY406 pKa = 7.36TGPSGIGPLDD416 pKa = 4.33DD417 pKa = 5.31EE418 pKa = 5.29NDD420 pKa = 3.62PSSAFIGIYY429 pKa = 10.17QYY431 pKa = 11.36DD432 pKa = 3.93ANNKK436 pKa = 9.17NILSGTVEE444 pKa = 4.1GSKK447 pKa = 10.86PEE449 pKa = 3.83
MM1 pKa = 7.16GAALGFSLVACGDD14 pKa = 4.02DD15 pKa = 4.23SDD17 pKa = 5.88SGDD20 pKa = 3.69DD21 pKa = 3.54TSGSDD26 pKa = 3.51EE27 pKa = 4.38SSSASADD34 pKa = 3.46TSEE37 pKa = 4.59SASEE41 pKa = 4.2SASADD46 pKa = 3.13AGGSFTNDD54 pKa = 2.31KK55 pKa = 10.72CAGSDD60 pKa = 3.08TSANKK65 pKa = 9.76FAVGGILPLTGNLAFLGPPAVAGVGLAVSDD95 pKa = 3.7INAAGGVNGAQACSNILDD113 pKa = 4.54SGDD116 pKa = 3.73STDD119 pKa = 3.8MSISTASAGTMVAAKK134 pKa = 9.2PSVVIGAEE142 pKa = 4.09SSSVSLNFVDD152 pKa = 4.02TLTDD156 pKa = 3.42AKK158 pKa = 9.61ITQISPANTAIDD170 pKa = 3.83LSGYY174 pKa = 9.48SPFYY178 pKa = 10.71FRR180 pKa = 11.84TAPPDD185 pKa = 3.84TVQGNALGTLISQDD199 pKa = 3.15GYY201 pKa = 9.09TNIGFLVFNDD211 pKa = 3.95TYY213 pKa = 11.22GTGLRR218 pKa = 11.84NAVQQTVEE226 pKa = 3.91AAGGKK231 pKa = 9.05CVYY234 pKa = 9.91GCKK237 pKa = 10.13GDD239 pKa = 4.08GDD241 pKa = 4.34EE242 pKa = 5.39FPAGQTTFSAEE253 pKa = 4.16VQAVTAAKK261 pKa = 9.51PDD263 pKa = 4.43AIVIIAFDD271 pKa = 3.8EE272 pKa = 4.54TKK274 pKa = 10.81SIVPEE279 pKa = 4.08LASSGFDD286 pKa = 3.01MSKK289 pKa = 9.61TYY291 pKa = 10.4FVDD294 pKa = 4.82GNLSDD299 pKa = 3.94YY300 pKa = 11.2GQDD303 pKa = 3.73FEE305 pKa = 5.7PGTLEE310 pKa = 4.3GAQGTLPGQDD320 pKa = 3.56PDD322 pKa = 4.72QGFKK326 pKa = 11.28DD327 pKa = 4.64NLTGWYY333 pKa = 9.79KK334 pKa = 10.48SANGEE339 pKa = 4.1DD340 pKa = 3.81LKK342 pKa = 11.15DD343 pKa = 3.54YY344 pKa = 11.1SYY346 pKa = 11.63GAEE349 pKa = 4.32SYY351 pKa = 10.87DD352 pKa = 3.59ATILAALAAAKK363 pKa = 10.41GGANDD368 pKa = 3.56SQTIQKK374 pKa = 9.87NLAAVSGATDD384 pKa = 3.78GEE386 pKa = 4.49EE387 pKa = 3.89CSTYY391 pKa = 11.22ADD393 pKa = 3.8CLKK396 pKa = 10.63LIEE399 pKa = 4.98GGSDD403 pKa = 2.54IHH405 pKa = 5.63YY406 pKa = 7.36TGPSGIGPLDD416 pKa = 4.33DD417 pKa = 5.31EE418 pKa = 5.29NDD420 pKa = 3.62PSSAFIGIYY429 pKa = 10.17QYY431 pKa = 11.36DD432 pKa = 3.93ANNKK436 pKa = 9.17NILSGTVEE444 pKa = 4.1GSKK447 pKa = 10.86PEE449 pKa = 3.83
Molecular weight: 45.34 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6G6W969|A0A6G6W969_9ACTN ROK family protein OS=Nocardioides anomalus OX=2712223 GN=G5V58_03015 PE=3 SV=1
MM1 pKa = 7.42ARR3 pKa = 11.84SHH5 pKa = 6.66LLKK8 pKa = 10.38ALQPARR14 pKa = 11.84VTGRR18 pKa = 11.84RR19 pKa = 11.84GRR21 pKa = 11.84GRR23 pKa = 11.84GRR25 pKa = 11.84GRR27 pKa = 11.84GHH29 pKa = 7.07TSTGTHH35 pKa = 6.43TGTGTRR41 pKa = 11.84TRR43 pKa = 11.84TRR45 pKa = 11.84THH47 pKa = 5.77GG48 pKa = 3.3
MM1 pKa = 7.42ARR3 pKa = 11.84SHH5 pKa = 6.66LLKK8 pKa = 10.38ALQPARR14 pKa = 11.84VTGRR18 pKa = 11.84RR19 pKa = 11.84GRR21 pKa = 11.84GRR23 pKa = 11.84GRR25 pKa = 11.84GRR27 pKa = 11.84GHH29 pKa = 7.07TSTGTHH35 pKa = 6.43TGTGTRR41 pKa = 11.84TRR43 pKa = 11.84TRR45 pKa = 11.84THH47 pKa = 5.77GG48 pKa = 3.3
Molecular weight: 5.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1544052 |
33 |
2111 |
314.2 |
33.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.478 ± 0.063 | 0.711 ± 0.009 |
6.56 ± 0.033 | 5.695 ± 0.037 |
2.729 ± 0.019 | 9.375 ± 0.037 |
2.218 ± 0.016 | 2.816 ± 0.025 |
1.756 ± 0.025 | 10.564 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.637 ± 0.014 | 1.57 ± 0.021 |
5.74 ± 0.03 | 2.856 ± 0.017 |
7.695 ± 0.037 | 5.119 ± 0.024 |
6.144 ± 0.036 | 9.789 ± 0.037 |
1.562 ± 0.015 | 1.989 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
