
Mastomys natalensis polyomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Betapolyomavirus
Average proteome isoelectric point is 5.87
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
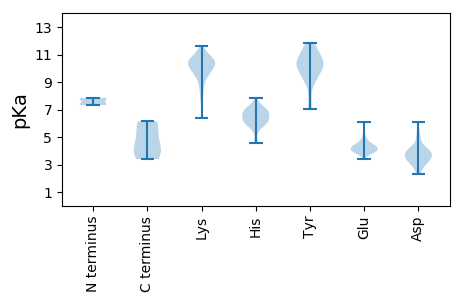
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|E5RUP2|E5RUP2_9POLY Minor capsid protein OS=Mastomys natalensis polyomavirus 1 OX=1891768 GN=VP3 PE=3 SV=1
MM1 pKa = 7.44GAFLTVLAEE10 pKa = 4.16VFEE13 pKa = 4.57LASVTGLSAEE23 pKa = 4.43SIISGEE29 pKa = 4.08AFTTAEE35 pKa = 4.49LLQSHH40 pKa = 7.17IANLVTYY47 pKa = 10.69GEE49 pKa = 4.21LTEE52 pKa = 5.18AEE54 pKa = 4.11ALAAAEE60 pKa = 4.43VSTEE64 pKa = 4.16AYY66 pKa = 9.86SALQTLTPNFPQVLAALAASEE87 pKa = 4.01ITATGSITVGAAVAAALYY105 pKa = 8.45PYY107 pKa = 9.68SYY109 pKa = 10.97EE110 pKa = 4.16FSTPLANLNQNMALQLWFPDD130 pKa = 2.57VDD132 pKa = 4.2YY133 pKa = 11.23NFPGLLPFVRR143 pKa = 11.84FVNYY147 pKa = 9.61IDD149 pKa = 3.84PARR152 pKa = 11.84WASDD156 pKa = 3.41LYY158 pKa = 10.78HH159 pKa = 6.97AVGRR163 pKa = 11.84YY164 pKa = 7.03FWQSVEE170 pKa = 4.25RR171 pKa = 11.84EE172 pKa = 3.9ASRR175 pKa = 11.84QIGDD179 pKa = 3.95LSRR182 pKa = 11.84DD183 pKa = 3.11LALRR187 pKa = 11.84TTQTVGEE194 pKa = 4.1TLARR198 pKa = 11.84YY199 pKa = 9.2FEE201 pKa = 4.06NARR204 pKa = 11.84WAVTNLPSLNLYY216 pKa = 10.41SGLEE220 pKa = 4.04TYY222 pKa = 10.24YY223 pKa = 11.39SKK225 pKa = 11.24LSPLNPIRR233 pKa = 11.84ARR235 pKa = 11.84QLARR239 pKa = 11.84NLGEE243 pKa = 4.19EE244 pKa = 3.83EE245 pKa = 4.31PYY247 pKa = 10.55RR248 pKa = 11.84YY249 pKa = 10.13DD250 pKa = 4.25RR251 pKa = 11.84YY252 pKa = 11.23DD253 pKa = 3.52GAQSNQKK260 pKa = 9.28SAEE263 pKa = 4.04YY264 pKa = 10.04VEE266 pKa = 5.84KK267 pKa = 10.82YY268 pKa = 10.23DD269 pKa = 4.16SPGGAHH275 pKa = 6.11QRR277 pKa = 11.84HH278 pKa = 5.59TPDD281 pKa = 2.5WMLPLILGLYY291 pKa = 10.37GDD293 pKa = 5.15ISPSWGEE300 pKa = 3.83TIKK303 pKa = 10.79EE304 pKa = 3.95IEE306 pKa = 4.3EE307 pKa = 4.3EE308 pKa = 3.84EE309 pKa = 4.48DD310 pKa = 3.74GPKK313 pKa = 10.16KK314 pKa = 10.21KK315 pKa = 10.28RR316 pKa = 11.84PRR318 pKa = 11.84KK319 pKa = 9.78AGWKK323 pKa = 9.73ASRR326 pKa = 11.84STKK329 pKa = 10.45ANHH332 pKa = 6.14
MM1 pKa = 7.44GAFLTVLAEE10 pKa = 4.16VFEE13 pKa = 4.57LASVTGLSAEE23 pKa = 4.43SIISGEE29 pKa = 4.08AFTTAEE35 pKa = 4.49LLQSHH40 pKa = 7.17IANLVTYY47 pKa = 10.69GEE49 pKa = 4.21LTEE52 pKa = 5.18AEE54 pKa = 4.11ALAAAEE60 pKa = 4.43VSTEE64 pKa = 4.16AYY66 pKa = 9.86SALQTLTPNFPQVLAALAASEE87 pKa = 4.01ITATGSITVGAAVAAALYY105 pKa = 8.45PYY107 pKa = 9.68SYY109 pKa = 10.97EE110 pKa = 4.16FSTPLANLNQNMALQLWFPDD130 pKa = 2.57VDD132 pKa = 4.2YY133 pKa = 11.23NFPGLLPFVRR143 pKa = 11.84FVNYY147 pKa = 9.61IDD149 pKa = 3.84PARR152 pKa = 11.84WASDD156 pKa = 3.41LYY158 pKa = 10.78HH159 pKa = 6.97AVGRR163 pKa = 11.84YY164 pKa = 7.03FWQSVEE170 pKa = 4.25RR171 pKa = 11.84EE172 pKa = 3.9ASRR175 pKa = 11.84QIGDD179 pKa = 3.95LSRR182 pKa = 11.84DD183 pKa = 3.11LALRR187 pKa = 11.84TTQTVGEE194 pKa = 4.1TLARR198 pKa = 11.84YY199 pKa = 9.2FEE201 pKa = 4.06NARR204 pKa = 11.84WAVTNLPSLNLYY216 pKa = 10.41SGLEE220 pKa = 4.04TYY222 pKa = 10.24YY223 pKa = 11.39SKK225 pKa = 11.24LSPLNPIRR233 pKa = 11.84ARR235 pKa = 11.84QLARR239 pKa = 11.84NLGEE243 pKa = 4.19EE244 pKa = 3.83EE245 pKa = 4.31PYY247 pKa = 10.55RR248 pKa = 11.84YY249 pKa = 10.13DD250 pKa = 4.25RR251 pKa = 11.84YY252 pKa = 11.23DD253 pKa = 3.52GAQSNQKK260 pKa = 9.28SAEE263 pKa = 4.04YY264 pKa = 10.04VEE266 pKa = 5.84KK267 pKa = 10.82YY268 pKa = 10.23DD269 pKa = 4.16SPGGAHH275 pKa = 6.11QRR277 pKa = 11.84HH278 pKa = 5.59TPDD281 pKa = 2.5WMLPLILGLYY291 pKa = 10.37GDD293 pKa = 5.15ISPSWGEE300 pKa = 3.83TIKK303 pKa = 10.79EE304 pKa = 3.95IEE306 pKa = 4.3EE307 pKa = 4.3EE308 pKa = 3.84EE309 pKa = 4.48DD310 pKa = 3.74GPKK313 pKa = 10.16KK314 pKa = 10.21KK315 pKa = 10.28RR316 pKa = 11.84PRR318 pKa = 11.84KK319 pKa = 9.78AGWKK323 pKa = 9.73ASRR326 pKa = 11.84STKK329 pKa = 10.45ANHH332 pKa = 6.14
Molecular weight: 36.84 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E5RUP1|E5RUP1_9POLY Minor capsid protein OS=Mastomys natalensis polyomavirus 1 OX=1891768 GN=VP2 PE=3 SV=1
MM1 pKa = 7.36IQVLWQTLKK10 pKa = 10.76CLLRR14 pKa = 11.84VFSISHH20 pKa = 6.69ASAAVLGDD28 pKa = 3.15IMCILEE34 pKa = 4.35EE35 pKa = 4.32FYY37 pKa = 11.48KK38 pKa = 10.61LAKK41 pKa = 8.58EE42 pKa = 4.28HH43 pKa = 6.16KK44 pKa = 7.9TVLNKK49 pKa = 9.35VEE51 pKa = 4.88CGWEE55 pKa = 3.74LARR58 pKa = 11.84FKK60 pKa = 11.16QKK62 pKa = 10.34INYY65 pKa = 9.43YY66 pKa = 7.29MDD68 pKa = 3.54KK69 pKa = 11.03VKK71 pKa = 10.12ATLKK75 pKa = 10.47HH76 pKa = 5.78CGKK79 pKa = 9.24VQVMYY84 pKa = 10.13FSKK87 pKa = 10.57QGTDD91 pKa = 3.08SVDD94 pKa = 3.3RR95 pKa = 11.84NLACVPEE102 pKa = 4.54LGKK105 pKa = 9.58GTLALSLGHH114 pKa = 6.01SEE116 pKa = 4.39QEE118 pKa = 4.29RR119 pKa = 11.84EE120 pKa = 4.03EE121 pKa = 4.25TACDD125 pKa = 3.65CLACQDD131 pKa = 3.96MRR133 pKa = 11.84QQLNLAEE140 pKa = 4.57HH141 pKa = 6.27LRR143 pKa = 11.84SLTEE147 pKa = 3.92EE148 pKa = 4.05LQKK151 pKa = 11.58LLMM154 pKa = 4.97
MM1 pKa = 7.36IQVLWQTLKK10 pKa = 10.76CLLRR14 pKa = 11.84VFSISHH20 pKa = 6.69ASAAVLGDD28 pKa = 3.15IMCILEE34 pKa = 4.35EE35 pKa = 4.32FYY37 pKa = 11.48KK38 pKa = 10.61LAKK41 pKa = 8.58EE42 pKa = 4.28HH43 pKa = 6.16KK44 pKa = 7.9TVLNKK49 pKa = 9.35VEE51 pKa = 4.88CGWEE55 pKa = 3.74LARR58 pKa = 11.84FKK60 pKa = 11.16QKK62 pKa = 10.34INYY65 pKa = 9.43YY66 pKa = 7.29MDD68 pKa = 3.54KK69 pKa = 11.03VKK71 pKa = 10.12ATLKK75 pKa = 10.47HH76 pKa = 5.78CGKK79 pKa = 9.24VQVMYY84 pKa = 10.13FSKK87 pKa = 10.57QGTDD91 pKa = 3.08SVDD94 pKa = 3.3RR95 pKa = 11.84NLACVPEE102 pKa = 4.54LGKK105 pKa = 9.58GTLALSLGHH114 pKa = 6.01SEE116 pKa = 4.39QEE118 pKa = 4.29RR119 pKa = 11.84EE120 pKa = 4.03EE121 pKa = 4.25TACDD125 pKa = 3.65CLACQDD131 pKa = 3.96MRR133 pKa = 11.84QQLNLAEE140 pKa = 4.57HH141 pKa = 6.27LRR143 pKa = 11.84SLTEE147 pKa = 3.92EE148 pKa = 4.05LQKK151 pKa = 11.58LLMM154 pKa = 4.97
Molecular weight: 17.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1876 |
154 |
653 |
312.7 |
35.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.716 ± 1.365 | 2.239 ± 0.728 |
5.171 ± 0.398 | 7.463 ± 0.308 |
3.945 ± 0.386 | 6.023 ± 0.907 |
1.866 ± 0.276 | 4.318 ± 0.382 |
6.503 ± 0.97 | 11.407 ± 0.748 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.452 ± 0.426 | 4.744 ± 0.428 |
5.277 ± 0.705 | 4.584 ± 0.304 |
4.904 ± 0.607 | 5.171 ± 0.698 |
5.97 ± 0.405 | 5.384 ± 0.548 |
1.599 ± 0.384 | 4.264 ± 0.508 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
