
Lachnospiraceae bacterium A10
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; unclassified Lachnospiraceae
Average proteome isoelectric point is 5.74
Get precalculated fractions of proteins
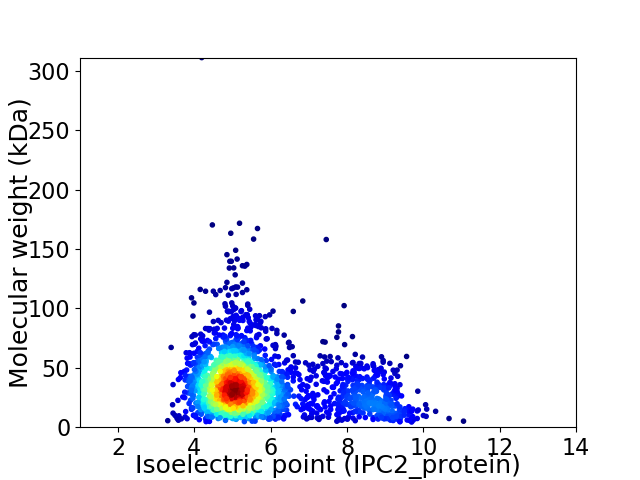
Virtual 2D-PAGE plot for 2244 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
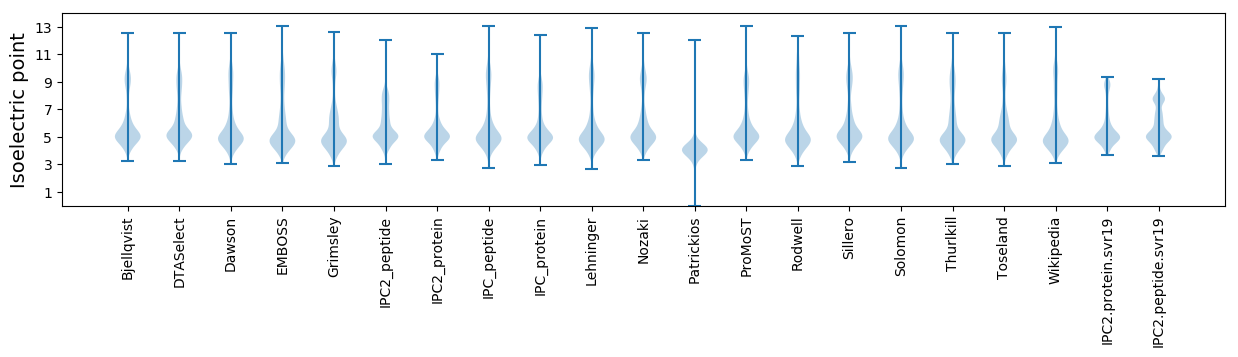
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H6UPR6|A0A1H6UPR6_9FIRM Uncharacterized protein OS=Lachnospiraceae bacterium A10 OX=1520821 GN=SAMN02910453_1645 PE=4 SV=1
MM1 pKa = 7.56KK2 pKa = 10.4FHH4 pKa = 7.11TNRR7 pKa = 11.84LQPKK11 pKa = 9.5RR12 pKa = 11.84FGALFLAASMLTLSACGSDD31 pKa = 3.81SISYY35 pKa = 10.27EE36 pKa = 3.97PLTEE40 pKa = 4.52EE41 pKa = 5.63DD42 pKa = 3.43DD43 pKa = 3.89TVYY46 pKa = 10.83NICIVQSEE54 pKa = 5.01DD55 pKa = 3.32NTEE58 pKa = 4.04LNQIQQGFQDD68 pKa = 3.85ALTDD72 pKa = 4.1LFGEE76 pKa = 4.16DD77 pKa = 3.58HH78 pKa = 7.17VNVTVQVANSEE89 pKa = 4.32TSVDD93 pKa = 4.74DD94 pKa = 3.76INSTYY99 pKa = 8.12TTDD102 pKa = 4.03EE103 pKa = 3.8NTQLIFANGSSALTSAAFYY122 pKa = 10.46TDD124 pKa = 5.22DD125 pKa = 4.12IPVVGAGVIDD135 pKa = 4.3FQSALHH141 pKa = 6.2TPDD144 pKa = 4.48SEE146 pKa = 4.05WDD148 pKa = 3.24AKK150 pKa = 10.46TGTNVTGISSRR161 pKa = 11.84PPIAAQLSLLIEE173 pKa = 4.47ATSDD177 pKa = 3.41LQTVGILYY185 pKa = 10.36CPEE188 pKa = 4.17DD189 pKa = 3.43TDD191 pKa = 5.9AIYY194 pKa = 10.8QNEE197 pKa = 4.1ILEE200 pKa = 5.05DD201 pKa = 3.79YY202 pKa = 10.78LDD204 pKa = 3.56QAGIAWRR211 pKa = 11.84EE212 pKa = 3.7YY213 pKa = 10.15EE214 pKa = 4.17IPSTEE219 pKa = 3.79AAITEE224 pKa = 4.52HH225 pKa = 6.1EE226 pKa = 4.26TSIDD230 pKa = 3.74TEE232 pKa = 4.64TSGSTISGDD241 pKa = 3.26KK242 pKa = 10.62VVANSAKK249 pKa = 9.71EE250 pKa = 3.79GSEE253 pKa = 3.57IDD255 pKa = 3.6VEE257 pKa = 5.17SIGEE261 pKa = 4.29SNVLFGLNSTNSARR275 pKa = 11.84SAKK278 pKa = 9.93ISANWTQDD286 pKa = 2.56INDD289 pKa = 4.11PVSYY293 pKa = 10.84DD294 pKa = 3.32LGSTTEE300 pKa = 4.21EE301 pKa = 3.89IVNYY305 pKa = 10.0AASQSSAFFISANSYY320 pKa = 11.19LDD322 pKa = 3.99DD323 pKa = 4.94QIDD326 pKa = 4.36TIASIANATGTTTVSADD343 pKa = 3.21TTIASKK349 pKa = 9.46TLVSLYY355 pKa = 9.85IDD357 pKa = 5.05PYY359 pKa = 10.66NEE361 pKa = 4.59GYY363 pKa = 10.19SAGKK367 pKa = 8.68LAYY370 pKa = 9.76RR371 pKa = 11.84ILVNGDD377 pKa = 3.44DD378 pKa = 4.25PGSIKK383 pKa = 9.45ITGTNADD390 pKa = 4.78DD391 pKa = 3.67ATKK394 pKa = 10.52LYY396 pKa = 10.46QDD398 pKa = 4.65TIAEE402 pKa = 4.23QFGLTFGKK410 pKa = 10.35SFSEE414 pKa = 4.13VDD416 pKa = 3.7DD417 pKa = 4.17FLNTYY422 pKa = 9.95VPGTNTEE429 pKa = 4.32RR430 pKa = 11.84ISSEE434 pKa = 3.94EE435 pKa = 3.87EE436 pKa = 3.63
MM1 pKa = 7.56KK2 pKa = 10.4FHH4 pKa = 7.11TNRR7 pKa = 11.84LQPKK11 pKa = 9.5RR12 pKa = 11.84FGALFLAASMLTLSACGSDD31 pKa = 3.81SISYY35 pKa = 10.27EE36 pKa = 3.97PLTEE40 pKa = 4.52EE41 pKa = 5.63DD42 pKa = 3.43DD43 pKa = 3.89TVYY46 pKa = 10.83NICIVQSEE54 pKa = 5.01DD55 pKa = 3.32NTEE58 pKa = 4.04LNQIQQGFQDD68 pKa = 3.85ALTDD72 pKa = 4.1LFGEE76 pKa = 4.16DD77 pKa = 3.58HH78 pKa = 7.17VNVTVQVANSEE89 pKa = 4.32TSVDD93 pKa = 4.74DD94 pKa = 3.76INSTYY99 pKa = 8.12TTDD102 pKa = 4.03EE103 pKa = 3.8NTQLIFANGSSALTSAAFYY122 pKa = 10.46TDD124 pKa = 5.22DD125 pKa = 4.12IPVVGAGVIDD135 pKa = 4.3FQSALHH141 pKa = 6.2TPDD144 pKa = 4.48SEE146 pKa = 4.05WDD148 pKa = 3.24AKK150 pKa = 10.46TGTNVTGISSRR161 pKa = 11.84PPIAAQLSLLIEE173 pKa = 4.47ATSDD177 pKa = 3.41LQTVGILYY185 pKa = 10.36CPEE188 pKa = 4.17DD189 pKa = 3.43TDD191 pKa = 5.9AIYY194 pKa = 10.8QNEE197 pKa = 4.1ILEE200 pKa = 5.05DD201 pKa = 3.79YY202 pKa = 10.78LDD204 pKa = 3.56QAGIAWRR211 pKa = 11.84EE212 pKa = 3.7YY213 pKa = 10.15EE214 pKa = 4.17IPSTEE219 pKa = 3.79AAITEE224 pKa = 4.52HH225 pKa = 6.1EE226 pKa = 4.26TSIDD230 pKa = 3.74TEE232 pKa = 4.64TSGSTISGDD241 pKa = 3.26KK242 pKa = 10.62VVANSAKK249 pKa = 9.71EE250 pKa = 3.79GSEE253 pKa = 3.57IDD255 pKa = 3.6VEE257 pKa = 5.17SIGEE261 pKa = 4.29SNVLFGLNSTNSARR275 pKa = 11.84SAKK278 pKa = 9.93ISANWTQDD286 pKa = 2.56INDD289 pKa = 4.11PVSYY293 pKa = 10.84DD294 pKa = 3.32LGSTTEE300 pKa = 4.21EE301 pKa = 3.89IVNYY305 pKa = 10.0AASQSSAFFISANSYY320 pKa = 11.19LDD322 pKa = 3.99DD323 pKa = 4.94QIDD326 pKa = 4.36TIASIANATGTTTVSADD343 pKa = 3.21TTIASKK349 pKa = 9.46TLVSLYY355 pKa = 9.85IDD357 pKa = 5.05PYY359 pKa = 10.66NEE361 pKa = 4.59GYY363 pKa = 10.19SAGKK367 pKa = 8.68LAYY370 pKa = 9.76RR371 pKa = 11.84ILVNGDD377 pKa = 3.44DD378 pKa = 4.25PGSIKK383 pKa = 9.45ITGTNADD390 pKa = 4.78DD391 pKa = 3.67ATKK394 pKa = 10.52LYY396 pKa = 10.46QDD398 pKa = 4.65TIAEE402 pKa = 4.23QFGLTFGKK410 pKa = 10.35SFSEE414 pKa = 4.13VDD416 pKa = 3.7DD417 pKa = 4.17FLNTYY422 pKa = 9.95VPGTNTEE429 pKa = 4.32RR430 pKa = 11.84ISSEE434 pKa = 3.94EE435 pKa = 3.87EE436 pKa = 3.63
Molecular weight: 46.9 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H6S7Y6|A0A1H6S7Y6_9FIRM Carbohydrate diacid regulator OS=Lachnospiraceae bacterium A10 OX=1520821 GN=SAMN02910453_0960 PE=4 SV=1
MM1 pKa = 7.6KK2 pKa = 8.55MTYY5 pKa = 8.41QPKK8 pKa = 8.94RR9 pKa = 11.84RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 8.92VHH16 pKa = 5.9GFRR19 pKa = 11.84ARR21 pKa = 11.84MATAGGRR28 pKa = 11.84KK29 pKa = 8.84VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.61GRR39 pKa = 11.84KK40 pKa = 8.87KK41 pKa = 10.59LSAA44 pKa = 3.95
MM1 pKa = 7.6KK2 pKa = 8.55MTYY5 pKa = 8.41QPKK8 pKa = 8.94RR9 pKa = 11.84RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 8.92VHH16 pKa = 5.9GFRR19 pKa = 11.84ARR21 pKa = 11.84MATAGGRR28 pKa = 11.84KK29 pKa = 8.84VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.61GRR39 pKa = 11.84KK40 pKa = 8.87KK41 pKa = 10.59LSAA44 pKa = 3.95
Molecular weight: 5.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
749661 |
37 |
2935 |
334.1 |
37.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.29 ± 0.049 | 1.298 ± 0.021 |
6.196 ± 0.041 | 7.876 ± 0.064 |
4.147 ± 0.037 | 6.741 ± 0.041 |
1.85 ± 0.021 | 7.501 ± 0.051 |
6.608 ± 0.047 | 8.74 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.323 ± 0.03 | 4.495 ± 0.037 |
3.003 ± 0.029 | 3.166 ± 0.026 |
4.198 ± 0.039 | 5.939 ± 0.052 |
5.446 ± 0.054 | 7.107 ± 0.044 |
0.818 ± 0.016 | 4.256 ± 0.039 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
