
Homoserinimonas aerilata
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Homoserinimonas
Average proteome isoelectric point is 5.94
Get precalculated fractions of proteins
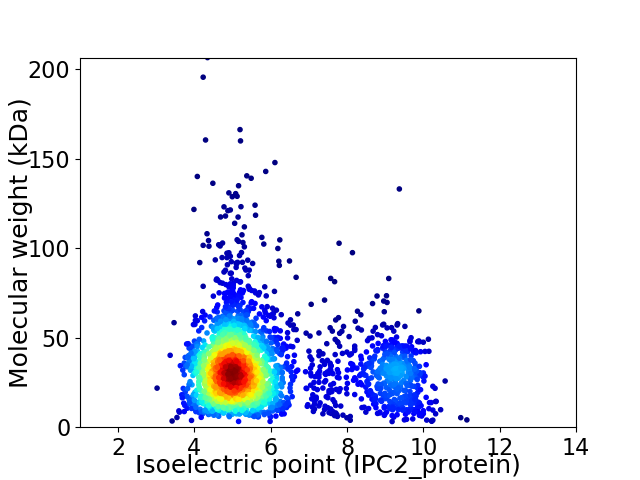
Virtual 2D-PAGE plot for 2633 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A542YHZ8|A0A542YHZ8_9MICO 2-hydroxychromene-2-carboxylate isomerase OS=Homoserinimonas aerilata OX=1162970 GN=FB562_0805 PE=4 SV=1
MM1 pKa = 7.92KK2 pKa = 10.21KK3 pKa = 10.43RR4 pKa = 11.84FMTSIAVGAVALLGLTACAGSADD27 pKa = 3.82TGDD30 pKa = 3.89NGSGDD35 pKa = 4.23GDD37 pKa = 4.05SKK39 pKa = 11.61SLTVGVFNGWPEE51 pKa = 4.14GEE53 pKa = 4.02AASYY57 pKa = 9.67LWKK60 pKa = 10.65AVLEE64 pKa = 4.25EE65 pKa = 3.93KK66 pKa = 10.76GYY68 pKa = 11.15DD69 pKa = 3.47VTLEE73 pKa = 4.02YY74 pKa = 11.15ADD76 pKa = 4.77AGPVFAGLSTGDD88 pKa = 3.42YY89 pKa = 11.18DD90 pKa = 4.34VAFDD94 pKa = 3.27GWLPLTHH101 pKa = 6.32QSYY104 pKa = 9.68MDD106 pKa = 3.64EE107 pKa = 4.57YY108 pKa = 11.2GDD110 pKa = 3.87SLEE113 pKa = 6.27DD114 pKa = 3.42LGSWNDD120 pKa = 3.72DD121 pKa = 3.02AVLTVAVNEE130 pKa = 4.38DD131 pKa = 3.56APITSLDD138 pKa = 3.68EE139 pKa = 4.55LADD142 pKa = 3.87HH143 pKa = 7.3ADD145 pKa = 3.39EE146 pKa = 4.7FGNRR150 pKa = 11.84IVGIEE155 pKa = 4.04PGAGLTKK162 pKa = 10.54AVGEE166 pKa = 4.21QTIPTYY172 pKa = 11.03GLDD175 pKa = 3.22GMEE178 pKa = 5.1FLTSSTPAMLSEE190 pKa = 4.83LSAALDD196 pKa = 3.59AGEE199 pKa = 4.25NVAVTLWRR207 pKa = 11.84PHH209 pKa = 5.02WAYY212 pKa = 10.82DD213 pKa = 3.5EE214 pKa = 4.31YY215 pKa = 10.64PIRR218 pKa = 11.84DD219 pKa = 3.72LEE221 pKa = 4.46DD222 pKa = 3.88PEE224 pKa = 4.53GTLGASEE231 pKa = 5.43GIHH234 pKa = 6.25SIARR238 pKa = 11.84SGFGDD243 pKa = 3.87EE244 pKa = 4.31FSEE247 pKa = 4.19LAGWIGDD254 pKa = 3.53WKK256 pKa = 10.79LDD258 pKa = 3.85SEE260 pKa = 4.79LLYY263 pKa = 11.06SLEE266 pKa = 3.71NAMYY270 pKa = 10.76NSGADD275 pKa = 3.22ASEE278 pKa = 4.47YY279 pKa = 10.79EE280 pKa = 4.79SIVADD285 pKa = 4.69WIAEE289 pKa = 3.93NQEE292 pKa = 4.25YY293 pKa = 11.1VDD295 pKa = 4.58GLTSS299 pKa = 3.2
MM1 pKa = 7.92KK2 pKa = 10.21KK3 pKa = 10.43RR4 pKa = 11.84FMTSIAVGAVALLGLTACAGSADD27 pKa = 3.82TGDD30 pKa = 3.89NGSGDD35 pKa = 4.23GDD37 pKa = 4.05SKK39 pKa = 11.61SLTVGVFNGWPEE51 pKa = 4.14GEE53 pKa = 4.02AASYY57 pKa = 9.67LWKK60 pKa = 10.65AVLEE64 pKa = 4.25EE65 pKa = 3.93KK66 pKa = 10.76GYY68 pKa = 11.15DD69 pKa = 3.47VTLEE73 pKa = 4.02YY74 pKa = 11.15ADD76 pKa = 4.77AGPVFAGLSTGDD88 pKa = 3.42YY89 pKa = 11.18DD90 pKa = 4.34VAFDD94 pKa = 3.27GWLPLTHH101 pKa = 6.32QSYY104 pKa = 9.68MDD106 pKa = 3.64EE107 pKa = 4.57YY108 pKa = 11.2GDD110 pKa = 3.87SLEE113 pKa = 6.27DD114 pKa = 3.42LGSWNDD120 pKa = 3.72DD121 pKa = 3.02AVLTVAVNEE130 pKa = 4.38DD131 pKa = 3.56APITSLDD138 pKa = 3.68EE139 pKa = 4.55LADD142 pKa = 3.87HH143 pKa = 7.3ADD145 pKa = 3.39EE146 pKa = 4.7FGNRR150 pKa = 11.84IVGIEE155 pKa = 4.04PGAGLTKK162 pKa = 10.54AVGEE166 pKa = 4.21QTIPTYY172 pKa = 11.03GLDD175 pKa = 3.22GMEE178 pKa = 5.1FLTSSTPAMLSEE190 pKa = 4.83LSAALDD196 pKa = 3.59AGEE199 pKa = 4.25NVAVTLWRR207 pKa = 11.84PHH209 pKa = 5.02WAYY212 pKa = 10.82DD213 pKa = 3.5EE214 pKa = 4.31YY215 pKa = 10.64PIRR218 pKa = 11.84DD219 pKa = 3.72LEE221 pKa = 4.46DD222 pKa = 3.88PEE224 pKa = 4.53GTLGASEE231 pKa = 5.43GIHH234 pKa = 6.25SIARR238 pKa = 11.84SGFGDD243 pKa = 3.87EE244 pKa = 4.31FSEE247 pKa = 4.19LAGWIGDD254 pKa = 3.53WKK256 pKa = 10.79LDD258 pKa = 3.85SEE260 pKa = 4.79LLYY263 pKa = 11.06SLEE266 pKa = 3.71NAMYY270 pKa = 10.76NSGADD275 pKa = 3.22ASEE278 pKa = 4.47YY279 pKa = 10.79EE280 pKa = 4.79SIVADD285 pKa = 4.69WIAEE289 pKa = 3.93NQEE292 pKa = 4.25YY293 pKa = 11.1VDD295 pKa = 4.58GLTSS299 pKa = 3.2
Molecular weight: 31.89 kDa
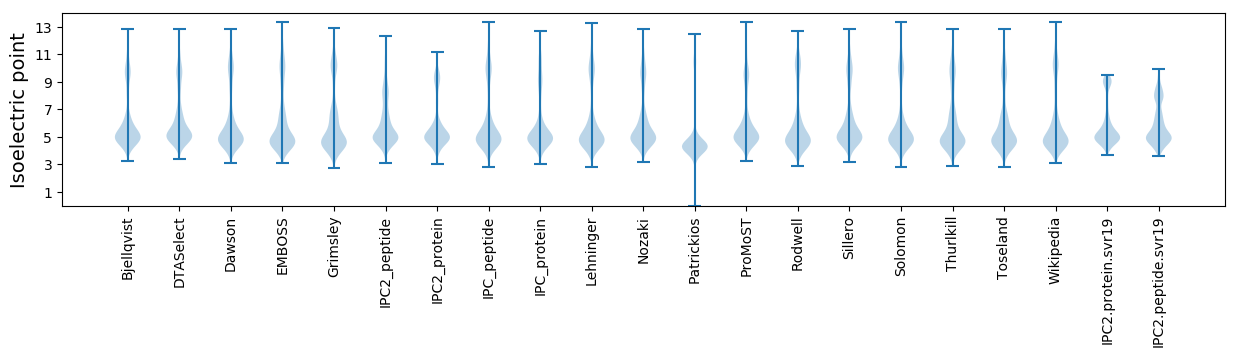
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A542YA47|A0A542YA47_9MICO HAD superfamily hydrolase (TIGR01490 family) OS=Homoserinimonas aerilata OX=1162970 GN=FB562_2380 PE=3 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
851979 |
30 |
1985 |
323.6 |
34.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.998 ± 0.074 | 0.476 ± 0.01 |
5.912 ± 0.041 | 5.818 ± 0.044 |
3.236 ± 0.028 | 8.989 ± 0.044 |
1.931 ± 0.023 | 4.918 ± 0.038 |
2.269 ± 0.036 | 10.295 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.906 ± 0.017 | 2.166 ± 0.026 |
5.118 ± 0.036 | 2.919 ± 0.027 |
6.776 ± 0.046 | 6.148 ± 0.034 |
5.947 ± 0.037 | 8.835 ± 0.044 |
1.4 ± 0.021 | 1.943 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
