
Oxalobacteraceae bacterium CAVE-383
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Oxalobacteraceae; unclassified Oxalobacteraceae
Average proteome isoelectric point is 6.74
Get precalculated fractions of proteins
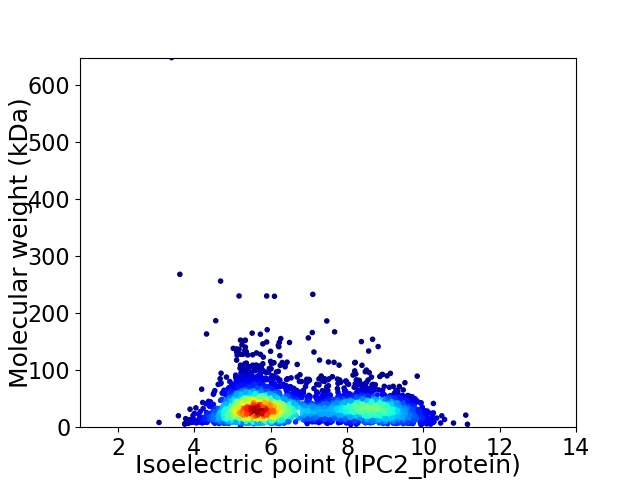
Virtual 2D-PAGE plot for 3601 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
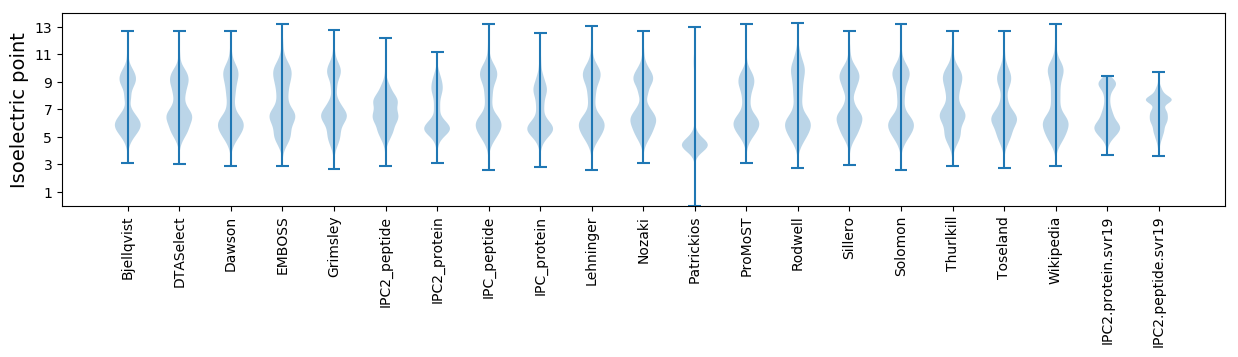
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4V1QP88|A0A4V1QP88_9BURK Uncharacterized protein OS=Oxalobacteraceae bacterium CAVE-383 OX=2478672 GN=D9O50_16250 PE=4 SV=1
MM1 pKa = 7.99PDD3 pKa = 3.09ANGDD7 pKa = 3.65FVAPVAKK14 pKa = 10.26SAIEE18 pKa = 3.88KK19 pKa = 9.2VQVVDD24 pKa = 3.26VDD26 pKa = 3.73MVLQLKK32 pKa = 10.46DD33 pKa = 3.42GTSMVLAGGAISAMDD48 pKa = 3.94ASHH51 pKa = 6.73TEE53 pKa = 4.04VKK55 pKa = 10.66FSDD58 pKa = 3.75GLQGLSKK65 pKa = 10.86LFDD68 pKa = 3.95LVGTIAIPKK77 pKa = 9.21LDD79 pKa = 5.54PILSSLQGHH88 pKa = 6.81PNSEE92 pKa = 4.23GQGAGANANQPAATEE107 pKa = 4.38SAGLTASVTSQIVNQLTQIVQANAQSVTIKK137 pKa = 10.05TVDD140 pKa = 3.46PSAKK144 pKa = 10.04VAVLSQSTKK153 pKa = 10.14TGDD156 pKa = 3.31VDD158 pKa = 4.27SQPLQHH164 pKa = 6.68TEE166 pKa = 3.75PPIPPEE172 pKa = 4.05RR173 pKa = 11.84PGVIPQEE180 pKa = 4.48HH181 pKa = 7.27IIGPDD186 pKa = 3.41APAMALSLVNLTTITQIGNVLYY208 pKa = 10.84GSGGTPASATDD219 pKa = 3.67AGNNAQFASQVINAPANATEE239 pKa = 3.89IHH241 pKa = 6.62AGANIPTNDD250 pKa = 3.85FIKK253 pKa = 10.95VVDD256 pKa = 3.52ITLTGNGIAQSIEE269 pKa = 4.05IKK271 pKa = 10.46GVPAGMSIANGTDD284 pKa = 2.73LGGGVWSIPVVSGQRR299 pKa = 11.84DD300 pKa = 3.3YY301 pKa = 11.54AIQIEE306 pKa = 5.01YY307 pKa = 7.51TTVPANPQTPIHH319 pKa = 5.6QQFSMEE325 pKa = 4.16FDD327 pKa = 3.5VTMTTADD334 pKa = 4.13GIVNLSQVKK343 pKa = 9.12QFVVKK348 pKa = 10.48DD349 pKa = 3.75VNSANDD355 pKa = 3.48LTFIDD360 pKa = 4.33STTGEE365 pKa = 4.33SVYY368 pKa = 10.87VLPAQGNSHH377 pKa = 6.54IVHH380 pKa = 6.81AGDD383 pKa = 4.06AGVTIIGSNANDD395 pKa = 3.67FLFGGAGADD404 pKa = 3.93TINGGTGNTYY414 pKa = 10.9LEE416 pKa = 4.67GGGGADD422 pKa = 3.76HH423 pKa = 7.24LNGGVGGINTAGYY436 pKa = 7.45TLSATGVTVDD446 pKa = 4.39LVAGTGTGGDD456 pKa = 3.64AQGDD460 pKa = 4.14TLTNIQNVIGSTHH473 pKa = 6.43NDD475 pKa = 3.12IIVASAAANNINGGGHH491 pKa = 6.9DD492 pKa = 4.75SIGGDD497 pKa = 3.41TVSYY501 pKa = 10.05AGSGTGVIIDD511 pKa = 4.34LAAGTGTGGNAAGDD525 pKa = 4.1KK526 pKa = 9.93LTGIQNVIGSDD537 pKa = 3.47HH538 pKa = 7.18NDD540 pKa = 3.01TFFGSSEE547 pKa = 3.84GNAFDD552 pKa = 5.82GGLHH556 pKa = 6.82DD557 pKa = 5.95LNGANTVSYY566 pKa = 11.49ANDD569 pKa = 3.56TSGTGAIVNLATGHH583 pKa = 6.25GSGAAAGDD591 pKa = 3.93SYY593 pKa = 12.19VNIQNIIGTAFNDD606 pKa = 3.84TFIDD610 pKa = 4.04GAGANRR616 pKa = 11.84YY617 pKa = 9.71DD618 pKa = 3.92GGVGGNDD625 pKa = 2.9TVDD628 pKa = 3.44YY629 pKa = 10.15SNAGAGVTVNFVTGHH644 pKa = 5.87GAGDD648 pKa = 3.42IAEE651 pKa = 4.72GDD653 pKa = 3.71TFVNVQNVIGSNFDD667 pKa = 3.73DD668 pKa = 4.35VFTAGIDD675 pKa = 3.59SKK677 pKa = 11.45NFNGGAGFDD686 pKa = 3.58TVRR689 pKa = 11.84YY690 pKa = 9.62DD691 pKa = 3.3SAASGIIIDD700 pKa = 5.32AINNVGRR707 pKa = 11.84GAAAGNTYY715 pKa = 9.99TGIEE719 pKa = 4.12AFVGGGSDD727 pKa = 4.4DD728 pKa = 3.81MFIASADD735 pKa = 3.53ANSFNGGAAGSDD747 pKa = 3.17TVDD750 pKa = 3.4YY751 pKa = 10.71SNSNAAVTVDD761 pKa = 3.59LFFTRR766 pKa = 11.84GQGTSGGYY774 pKa = 9.59AQGDD778 pKa = 3.9VLVNIANVIGSAGDD792 pKa = 3.93DD793 pKa = 3.38LFLANSVQNSFEE805 pKa = 4.25GGGGNNTVSYY815 pKa = 9.08EE816 pKa = 4.11HH817 pKa = 7.01SSSADD822 pKa = 3.29STGVIVDD829 pKa = 4.43LSATDD834 pKa = 4.09GSGTSGNYY842 pKa = 10.37ALGDD846 pKa = 3.56KK847 pKa = 10.41FSNIQNVIGSAFNDD861 pKa = 3.61TFVASSVANVFTGGGGIDD879 pKa = 3.3TVSYY883 pKa = 10.93AKK885 pKa = 9.38STSADD890 pKa = 2.89GVTGVTVDD898 pKa = 3.98LSTVDD903 pKa = 3.54GLGGAGHH910 pKa = 6.28GHH912 pKa = 6.3GNYY915 pKa = 10.31AEE917 pKa = 5.05GDD919 pKa = 3.87TIINVQNAIGSIYY932 pKa = 10.42NDD934 pKa = 3.62TFIGGAVANTFTGGGGSDD952 pKa = 3.24TVSYY956 pKa = 10.73AGSTAVTVDD965 pKa = 3.53LVNGVGVGGNAEE977 pKa = 3.76GDD979 pKa = 3.59KK980 pKa = 10.83YY981 pKa = 11.43INVQNVIGGDD991 pKa = 3.6ADD993 pKa = 3.73DD994 pKa = 4.96TIIDD998 pKa = 3.7NLAANTIDD1006 pKa = 3.67GGTVSLHH1013 pKa = 5.73NRR1015 pKa = 11.84VSYY1018 pKa = 11.03ANSVSSPGVAAGVTVDD1034 pKa = 3.87LTATDD1039 pKa = 3.9GTGTSGGYY1047 pKa = 8.32ATGDD1051 pKa = 3.44KK1052 pKa = 10.67LSNIQDD1058 pKa = 3.95LTGSAADD1065 pKa = 3.74DD1066 pKa = 3.78TFVASLAANNLDD1078 pKa = 4.01GGASTAASHH1087 pKa = 6.53NRR1089 pKa = 11.84VSYY1092 pKa = 10.64DD1093 pKa = 3.05KK1094 pKa = 11.33SVDD1097 pKa = 3.36GSGAAAGVTVDD1108 pKa = 4.14LVNGVGTGGYY1118 pKa = 9.85AQGDD1122 pKa = 4.23TYY1124 pKa = 12.03ANIQDD1129 pKa = 3.94VTGSKK1134 pKa = 10.71ADD1136 pKa = 3.85DD1137 pKa = 4.89LIIDD1141 pKa = 4.02SAAANTIDD1149 pKa = 3.89GGTGSLHH1156 pKa = 5.46NTVSYY1161 pKa = 11.04ANSVTSTGAAAGVTINLNLADD1182 pKa = 4.09GSGNSGGYY1190 pKa = 10.1AVGDD1194 pKa = 3.6KK1195 pKa = 10.84LSNIQNLVGSQADD1208 pKa = 3.68DD1209 pKa = 3.69TFVASAAANFLDD1221 pKa = 4.29GGTGSLHH1228 pKa = 6.2NRR1230 pKa = 11.84VDD1232 pKa = 3.65YY1233 pKa = 10.82SASVDD1238 pKa = 3.44AGNAAAAVTVDD1249 pKa = 3.51LSGNTGTGGYY1259 pKa = 9.86AQGDD1263 pKa = 4.23TYY1265 pKa = 12.03ANIQDD1270 pKa = 3.99VTGSGADD1277 pKa = 3.58DD1278 pKa = 4.44LFIASTAANNFDD1290 pKa = 4.1GGVSTAASHH1299 pKa = 6.26NRR1301 pKa = 11.84VSYY1304 pKa = 9.14SQSGTGVVVDD1314 pKa = 5.11LSVTNGNGTSGGAAGDD1330 pKa = 3.81TFVNIQDD1337 pKa = 3.54ITGSNQNDD1345 pKa = 3.68TFVASTAANNFDD1357 pKa = 4.2GGTGSDD1363 pKa = 3.23TVSYY1367 pKa = 11.16ARR1369 pKa = 11.84VTGGVGVTVDD1379 pKa = 3.79LTAGGALTGGAAAGDD1394 pKa = 4.25TYY1396 pKa = 11.4TSIEE1400 pKa = 4.1NVIGSANNDD1409 pKa = 3.42LFIDD1413 pKa = 4.27GGGAVPNAYY1422 pKa = 9.04TGGGGVDD1429 pKa = 3.98RR1430 pKa = 11.84ISYY1433 pKa = 9.98DD1434 pKa = 3.13NSAAGVNVDD1443 pKa = 5.32FGTGLNMGTGTGSYY1457 pKa = 10.27AQGDD1461 pKa = 4.55TYY1463 pKa = 11.68VGIQNVTGSKK1473 pKa = 10.41FDD1475 pKa = 4.8DD1476 pKa = 3.83NFTASTTVSAATANAFDD1493 pKa = 4.71GNTGTLHH1500 pKa = 6.6NRR1502 pKa = 11.84VLYY1505 pKa = 9.57TGTGTGVTVNLSNIIVNGVAAGSGGGGAAGDD1536 pKa = 4.07TYY1538 pKa = 11.97ANIQDD1543 pKa = 3.64ITGTTSDD1550 pKa = 3.94DD1551 pKa = 4.06RR1552 pKa = 11.84IIASAAANNIDD1563 pKa = 3.84GGTGSLHH1570 pKa = 6.23NRR1572 pKa = 11.84VDD1574 pKa = 3.61YY1575 pKa = 8.84TASVDD1580 pKa = 3.98AGNAPAGVTVDD1591 pKa = 3.81LSGNTGTGGYY1601 pKa = 9.86AQGDD1605 pKa = 4.23TYY1607 pKa = 12.03ANIQDD1612 pKa = 3.99VTGSGADD1619 pKa = 3.54DD1620 pKa = 4.48LFIASAAANNFDD1632 pKa = 4.37GGVSTAASHH1641 pKa = 6.35NRR1643 pKa = 11.84VSYY1646 pKa = 9.8LQSATGVVVDD1656 pKa = 5.88LSVTDD1661 pKa = 3.86GTGTSGGAAGDD1672 pKa = 3.81TFVNIQDD1679 pKa = 3.54ITGSNQNDD1687 pKa = 3.68TFVASTAANNFDD1699 pKa = 4.2GGTGSDD1705 pKa = 3.23TVSYY1709 pKa = 11.16ARR1711 pKa = 11.84VTGGVGVTVDD1721 pKa = 3.79LTAGGALTGGAAAGDD1736 pKa = 4.25TYY1738 pKa = 11.4TSIEE1742 pKa = 4.1NVIGSANNDD1751 pKa = 3.42LFIDD1755 pKa = 4.27GGGAVPNAYY1764 pKa = 9.04TGGGGVDD1771 pKa = 3.98RR1772 pKa = 11.84ISYY1775 pKa = 9.98DD1776 pKa = 3.13NSAAGVNVDD1785 pKa = 5.32FGTGLNMGTGTGSYY1799 pKa = 10.27AQGDD1803 pKa = 4.55TYY1805 pKa = 11.68VGIQNVTGSKK1815 pKa = 10.41FDD1817 pKa = 4.8DD1818 pKa = 3.83NFTASTTVSAATANAFDD1835 pKa = 4.71GNTGTLHH1842 pKa = 6.6NRR1844 pKa = 11.84VLYY1847 pKa = 9.57TGTGTGVTVNLSNIIVNGVAAGSGGGGAAGDD1878 pKa = 4.07TYY1880 pKa = 11.97ANIQDD1885 pKa = 3.63ITGTASDD1892 pKa = 3.94DD1893 pKa = 4.08TIVASSAANNIDD1905 pKa = 3.59GGASTAASHH1914 pKa = 6.51NRR1916 pKa = 11.84VDD1918 pKa = 3.81YY1919 pKa = 10.74SASVDD1924 pKa = 3.41AGNVAVGVTVDD1935 pKa = 4.22LLNSVGTGGYY1945 pKa = 9.91AQGDD1949 pKa = 4.23TYY1951 pKa = 12.03ANIQDD1956 pKa = 3.87VTGSGAADD1964 pKa = 3.44TFVASAAANKK1974 pKa = 9.8FDD1976 pKa = 4.37GGAGSDD1982 pKa = 3.46TVDD1985 pKa = 3.43YY1986 pKa = 9.08STSTAIIVNLSALTQNTLAAGTGSGGLAAGDD2017 pKa = 3.55TYY2019 pKa = 11.62TNIEE2023 pKa = 4.31NIKK2026 pKa = 10.44GGAGNDD2032 pKa = 3.23LFYY2035 pKa = 11.24AGTAANAFTGGGGIDD2050 pKa = 3.37TVSYY2054 pKa = 10.7VSSTNGVTASLIANAVGSGGDD2075 pKa = 3.34AAGDD2079 pKa = 3.73TFSGITNLTGSAQNDD2094 pKa = 4.13TLTGLAAGGSTLNGGAGSDD2113 pKa = 3.72TLNATGDD2120 pKa = 3.55NNIFIGGAGGDD2131 pKa = 3.56TMTGNGGLNNTADD2144 pKa = 3.99YY2145 pKa = 8.94TGSGAVVVDD2154 pKa = 4.7LFFTDD2159 pKa = 3.84GTGTSGGDD2167 pKa = 3.18ATGDD2171 pKa = 3.54KK2172 pKa = 10.91LSGIQNVKK2180 pKa = 10.54GSGGNDD2186 pKa = 2.75TFFANSVANVFTGNGGTDD2204 pKa = 3.22TVSYY2208 pKa = 10.21SHH2210 pKa = 6.41STTTVIASLLAGVGGTSGDD2229 pKa = 3.71ATGDD2233 pKa = 3.45TYY2235 pKa = 11.84NAITNLTGGSGDD2247 pKa = 3.71DD2248 pKa = 3.55TLTGLLAGGSTLVGNAGNDD2267 pKa = 3.21RR2268 pKa = 11.84LIATGGNNVLMGGAGADD2285 pKa = 3.39NMSATGANNTASYY2298 pKa = 10.77AGSTNGNIIVDD2309 pKa = 4.08LTKK2312 pKa = 10.46TDD2314 pKa = 3.48GTGTSGGDD2322 pKa = 3.13AAGDD2326 pKa = 3.51ILSGIQNVIGGLGDD2340 pKa = 3.87DD2341 pKa = 5.44KK2342 pKa = 11.46FILDD2346 pKa = 4.01HH2347 pKa = 6.38NANIIDD2353 pKa = 4.39GGNGSNTVSYY2363 pKa = 10.38EE2364 pKa = 3.75HH2365 pKa = 6.86EE2366 pKa = 4.04TTINVTVDD2374 pKa = 3.23LSGTGIANGDD2384 pKa = 4.08GLFGAQDD2391 pKa = 3.71TLTNIQNAIGGGGNDD2406 pKa = 3.55IFLAGNAHH2414 pKa = 6.11NFFDD2418 pKa = 4.65GGGGVDD2424 pKa = 3.42TVSYY2428 pKa = 11.06ANNNGAHH2435 pKa = 6.39GAAIAANTAGVTVDD2449 pKa = 4.01FHH2451 pKa = 8.68GNIGSGNWAAGDD2463 pKa = 3.57TYY2465 pKa = 11.41TNIEE2469 pKa = 4.18NVIGTTGNDD2478 pKa = 3.28TFIDD2482 pKa = 3.89AGDD2485 pKa = 4.12GVHH2488 pKa = 7.04NIYY2491 pKa = 10.97NGGANGAGSNDD2502 pKa = 3.25TVSYY2506 pKa = 10.51QFSATGGALTLSIDD2520 pKa = 3.68GTHH2523 pKa = 6.83ANAGNAAGDD2532 pKa = 3.79VFTNIANLTGSDD2544 pKa = 3.65TVNSEE2549 pKa = 3.55LWGDD2553 pKa = 3.65GNANILTAMGSTNTNILHH2571 pKa = 6.24GNGGGAGTDD2580 pKa = 3.47VLDD2583 pKa = 4.56GRR2585 pKa = 11.84LGGHH2589 pKa = 5.58NTLIAGDD2596 pKa = 3.93TGSTTVDD2603 pKa = 2.95INASADD2609 pKa = 3.51PNHH2612 pKa = 7.02AGGSILGTNIDD2623 pKa = 4.02QIVGSTTGAVTVKK2636 pKa = 9.84MFNLTTALDD2645 pKa = 3.52ISSFSATATVGGPKK2659 pKa = 9.71VSHH2662 pKa = 6.31ITTLDD2667 pKa = 3.38LSGDD2671 pKa = 3.79GVGTQISISANDD2683 pKa = 3.79VIALGNNHH2691 pKa = 5.69QLSVRR2696 pKa = 11.84MDD2698 pKa = 3.69GNDD2701 pKa = 3.85SINLVLQAGEE2711 pKa = 4.02HH2712 pKa = 6.03SVLFGNGDD2720 pKa = 3.66YY2721 pKa = 11.57AFFDD2725 pKa = 3.99ANNVEE2730 pKa = 4.59LARR2733 pKa = 11.84IHH2735 pKa = 6.33QLTT2738 pKa = 3.51
MM1 pKa = 7.99PDD3 pKa = 3.09ANGDD7 pKa = 3.65FVAPVAKK14 pKa = 10.26SAIEE18 pKa = 3.88KK19 pKa = 9.2VQVVDD24 pKa = 3.26VDD26 pKa = 3.73MVLQLKK32 pKa = 10.46DD33 pKa = 3.42GTSMVLAGGAISAMDD48 pKa = 3.94ASHH51 pKa = 6.73TEE53 pKa = 4.04VKK55 pKa = 10.66FSDD58 pKa = 3.75GLQGLSKK65 pKa = 10.86LFDD68 pKa = 3.95LVGTIAIPKK77 pKa = 9.21LDD79 pKa = 5.54PILSSLQGHH88 pKa = 6.81PNSEE92 pKa = 4.23GQGAGANANQPAATEE107 pKa = 4.38SAGLTASVTSQIVNQLTQIVQANAQSVTIKK137 pKa = 10.05TVDD140 pKa = 3.46PSAKK144 pKa = 10.04VAVLSQSTKK153 pKa = 10.14TGDD156 pKa = 3.31VDD158 pKa = 4.27SQPLQHH164 pKa = 6.68TEE166 pKa = 3.75PPIPPEE172 pKa = 4.05RR173 pKa = 11.84PGVIPQEE180 pKa = 4.48HH181 pKa = 7.27IIGPDD186 pKa = 3.41APAMALSLVNLTTITQIGNVLYY208 pKa = 10.84GSGGTPASATDD219 pKa = 3.67AGNNAQFASQVINAPANATEE239 pKa = 3.89IHH241 pKa = 6.62AGANIPTNDD250 pKa = 3.85FIKK253 pKa = 10.95VVDD256 pKa = 3.52ITLTGNGIAQSIEE269 pKa = 4.05IKK271 pKa = 10.46GVPAGMSIANGTDD284 pKa = 2.73LGGGVWSIPVVSGQRR299 pKa = 11.84DD300 pKa = 3.3YY301 pKa = 11.54AIQIEE306 pKa = 5.01YY307 pKa = 7.51TTVPANPQTPIHH319 pKa = 5.6QQFSMEE325 pKa = 4.16FDD327 pKa = 3.5VTMTTADD334 pKa = 4.13GIVNLSQVKK343 pKa = 9.12QFVVKK348 pKa = 10.48DD349 pKa = 3.75VNSANDD355 pKa = 3.48LTFIDD360 pKa = 4.33STTGEE365 pKa = 4.33SVYY368 pKa = 10.87VLPAQGNSHH377 pKa = 6.54IVHH380 pKa = 6.81AGDD383 pKa = 4.06AGVTIIGSNANDD395 pKa = 3.67FLFGGAGADD404 pKa = 3.93TINGGTGNTYY414 pKa = 10.9LEE416 pKa = 4.67GGGGADD422 pKa = 3.76HH423 pKa = 7.24LNGGVGGINTAGYY436 pKa = 7.45TLSATGVTVDD446 pKa = 4.39LVAGTGTGGDD456 pKa = 3.64AQGDD460 pKa = 4.14TLTNIQNVIGSTHH473 pKa = 6.43NDD475 pKa = 3.12IIVASAAANNINGGGHH491 pKa = 6.9DD492 pKa = 4.75SIGGDD497 pKa = 3.41TVSYY501 pKa = 10.05AGSGTGVIIDD511 pKa = 4.34LAAGTGTGGNAAGDD525 pKa = 4.1KK526 pKa = 9.93LTGIQNVIGSDD537 pKa = 3.47HH538 pKa = 7.18NDD540 pKa = 3.01TFFGSSEE547 pKa = 3.84GNAFDD552 pKa = 5.82GGLHH556 pKa = 6.82DD557 pKa = 5.95LNGANTVSYY566 pKa = 11.49ANDD569 pKa = 3.56TSGTGAIVNLATGHH583 pKa = 6.25GSGAAAGDD591 pKa = 3.93SYY593 pKa = 12.19VNIQNIIGTAFNDD606 pKa = 3.84TFIDD610 pKa = 4.04GAGANRR616 pKa = 11.84YY617 pKa = 9.71DD618 pKa = 3.92GGVGGNDD625 pKa = 2.9TVDD628 pKa = 3.44YY629 pKa = 10.15SNAGAGVTVNFVTGHH644 pKa = 5.87GAGDD648 pKa = 3.42IAEE651 pKa = 4.72GDD653 pKa = 3.71TFVNVQNVIGSNFDD667 pKa = 3.73DD668 pKa = 4.35VFTAGIDD675 pKa = 3.59SKK677 pKa = 11.45NFNGGAGFDD686 pKa = 3.58TVRR689 pKa = 11.84YY690 pKa = 9.62DD691 pKa = 3.3SAASGIIIDD700 pKa = 5.32AINNVGRR707 pKa = 11.84GAAAGNTYY715 pKa = 9.99TGIEE719 pKa = 4.12AFVGGGSDD727 pKa = 4.4DD728 pKa = 3.81MFIASADD735 pKa = 3.53ANSFNGGAAGSDD747 pKa = 3.17TVDD750 pKa = 3.4YY751 pKa = 10.71SNSNAAVTVDD761 pKa = 3.59LFFTRR766 pKa = 11.84GQGTSGGYY774 pKa = 9.59AQGDD778 pKa = 3.9VLVNIANVIGSAGDD792 pKa = 3.93DD793 pKa = 3.38LFLANSVQNSFEE805 pKa = 4.25GGGGNNTVSYY815 pKa = 9.08EE816 pKa = 4.11HH817 pKa = 7.01SSSADD822 pKa = 3.29STGVIVDD829 pKa = 4.43LSATDD834 pKa = 4.09GSGTSGNYY842 pKa = 10.37ALGDD846 pKa = 3.56KK847 pKa = 10.41FSNIQNVIGSAFNDD861 pKa = 3.61TFVASSVANVFTGGGGIDD879 pKa = 3.3TVSYY883 pKa = 10.93AKK885 pKa = 9.38STSADD890 pKa = 2.89GVTGVTVDD898 pKa = 3.98LSTVDD903 pKa = 3.54GLGGAGHH910 pKa = 6.28GHH912 pKa = 6.3GNYY915 pKa = 10.31AEE917 pKa = 5.05GDD919 pKa = 3.87TIINVQNAIGSIYY932 pKa = 10.42NDD934 pKa = 3.62TFIGGAVANTFTGGGGSDD952 pKa = 3.24TVSYY956 pKa = 10.73AGSTAVTVDD965 pKa = 3.53LVNGVGVGGNAEE977 pKa = 3.76GDD979 pKa = 3.59KK980 pKa = 10.83YY981 pKa = 11.43INVQNVIGGDD991 pKa = 3.6ADD993 pKa = 3.73DD994 pKa = 4.96TIIDD998 pKa = 3.7NLAANTIDD1006 pKa = 3.67GGTVSLHH1013 pKa = 5.73NRR1015 pKa = 11.84VSYY1018 pKa = 11.03ANSVSSPGVAAGVTVDD1034 pKa = 3.87LTATDD1039 pKa = 3.9GTGTSGGYY1047 pKa = 8.32ATGDD1051 pKa = 3.44KK1052 pKa = 10.67LSNIQDD1058 pKa = 3.95LTGSAADD1065 pKa = 3.74DD1066 pKa = 3.78TFVASLAANNLDD1078 pKa = 4.01GGASTAASHH1087 pKa = 6.53NRR1089 pKa = 11.84VSYY1092 pKa = 10.64DD1093 pKa = 3.05KK1094 pKa = 11.33SVDD1097 pKa = 3.36GSGAAAGVTVDD1108 pKa = 4.14LVNGVGTGGYY1118 pKa = 9.85AQGDD1122 pKa = 4.23TYY1124 pKa = 12.03ANIQDD1129 pKa = 3.94VTGSKK1134 pKa = 10.71ADD1136 pKa = 3.85DD1137 pKa = 4.89LIIDD1141 pKa = 4.02SAAANTIDD1149 pKa = 3.89GGTGSLHH1156 pKa = 5.46NTVSYY1161 pKa = 11.04ANSVTSTGAAAGVTINLNLADD1182 pKa = 4.09GSGNSGGYY1190 pKa = 10.1AVGDD1194 pKa = 3.6KK1195 pKa = 10.84LSNIQNLVGSQADD1208 pKa = 3.68DD1209 pKa = 3.69TFVASAAANFLDD1221 pKa = 4.29GGTGSLHH1228 pKa = 6.2NRR1230 pKa = 11.84VDD1232 pKa = 3.65YY1233 pKa = 10.82SASVDD1238 pKa = 3.44AGNAAAAVTVDD1249 pKa = 3.51LSGNTGTGGYY1259 pKa = 9.86AQGDD1263 pKa = 4.23TYY1265 pKa = 12.03ANIQDD1270 pKa = 3.99VTGSGADD1277 pKa = 3.58DD1278 pKa = 4.44LFIASTAANNFDD1290 pKa = 4.1GGVSTAASHH1299 pKa = 6.26NRR1301 pKa = 11.84VSYY1304 pKa = 9.14SQSGTGVVVDD1314 pKa = 5.11LSVTNGNGTSGGAAGDD1330 pKa = 3.81TFVNIQDD1337 pKa = 3.54ITGSNQNDD1345 pKa = 3.68TFVASTAANNFDD1357 pKa = 4.2GGTGSDD1363 pKa = 3.23TVSYY1367 pKa = 11.16ARR1369 pKa = 11.84VTGGVGVTVDD1379 pKa = 3.79LTAGGALTGGAAAGDD1394 pKa = 4.25TYY1396 pKa = 11.4TSIEE1400 pKa = 4.1NVIGSANNDD1409 pKa = 3.42LFIDD1413 pKa = 4.27GGGAVPNAYY1422 pKa = 9.04TGGGGVDD1429 pKa = 3.98RR1430 pKa = 11.84ISYY1433 pKa = 9.98DD1434 pKa = 3.13NSAAGVNVDD1443 pKa = 5.32FGTGLNMGTGTGSYY1457 pKa = 10.27AQGDD1461 pKa = 4.55TYY1463 pKa = 11.68VGIQNVTGSKK1473 pKa = 10.41FDD1475 pKa = 4.8DD1476 pKa = 3.83NFTASTTVSAATANAFDD1493 pKa = 4.71GNTGTLHH1500 pKa = 6.6NRR1502 pKa = 11.84VLYY1505 pKa = 9.57TGTGTGVTVNLSNIIVNGVAAGSGGGGAAGDD1536 pKa = 4.07TYY1538 pKa = 11.97ANIQDD1543 pKa = 3.64ITGTTSDD1550 pKa = 3.94DD1551 pKa = 4.06RR1552 pKa = 11.84IIASAAANNIDD1563 pKa = 3.84GGTGSLHH1570 pKa = 6.23NRR1572 pKa = 11.84VDD1574 pKa = 3.61YY1575 pKa = 8.84TASVDD1580 pKa = 3.98AGNAPAGVTVDD1591 pKa = 3.81LSGNTGTGGYY1601 pKa = 9.86AQGDD1605 pKa = 4.23TYY1607 pKa = 12.03ANIQDD1612 pKa = 3.99VTGSGADD1619 pKa = 3.54DD1620 pKa = 4.48LFIASAAANNFDD1632 pKa = 4.37GGVSTAASHH1641 pKa = 6.35NRR1643 pKa = 11.84VSYY1646 pKa = 9.8LQSATGVVVDD1656 pKa = 5.88LSVTDD1661 pKa = 3.86GTGTSGGAAGDD1672 pKa = 3.81TFVNIQDD1679 pKa = 3.54ITGSNQNDD1687 pKa = 3.68TFVASTAANNFDD1699 pKa = 4.2GGTGSDD1705 pKa = 3.23TVSYY1709 pKa = 11.16ARR1711 pKa = 11.84VTGGVGVTVDD1721 pKa = 3.79LTAGGALTGGAAAGDD1736 pKa = 4.25TYY1738 pKa = 11.4TSIEE1742 pKa = 4.1NVIGSANNDD1751 pKa = 3.42LFIDD1755 pKa = 4.27GGGAVPNAYY1764 pKa = 9.04TGGGGVDD1771 pKa = 3.98RR1772 pKa = 11.84ISYY1775 pKa = 9.98DD1776 pKa = 3.13NSAAGVNVDD1785 pKa = 5.32FGTGLNMGTGTGSYY1799 pKa = 10.27AQGDD1803 pKa = 4.55TYY1805 pKa = 11.68VGIQNVTGSKK1815 pKa = 10.41FDD1817 pKa = 4.8DD1818 pKa = 3.83NFTASTTVSAATANAFDD1835 pKa = 4.71GNTGTLHH1842 pKa = 6.6NRR1844 pKa = 11.84VLYY1847 pKa = 9.57TGTGTGVTVNLSNIIVNGVAAGSGGGGAAGDD1878 pKa = 4.07TYY1880 pKa = 11.97ANIQDD1885 pKa = 3.63ITGTASDD1892 pKa = 3.94DD1893 pKa = 4.08TIVASSAANNIDD1905 pKa = 3.59GGASTAASHH1914 pKa = 6.51NRR1916 pKa = 11.84VDD1918 pKa = 3.81YY1919 pKa = 10.74SASVDD1924 pKa = 3.41AGNVAVGVTVDD1935 pKa = 4.22LLNSVGTGGYY1945 pKa = 9.91AQGDD1949 pKa = 4.23TYY1951 pKa = 12.03ANIQDD1956 pKa = 3.87VTGSGAADD1964 pKa = 3.44TFVASAAANKK1974 pKa = 9.8FDD1976 pKa = 4.37GGAGSDD1982 pKa = 3.46TVDD1985 pKa = 3.43YY1986 pKa = 9.08STSTAIIVNLSALTQNTLAAGTGSGGLAAGDD2017 pKa = 3.55TYY2019 pKa = 11.62TNIEE2023 pKa = 4.31NIKK2026 pKa = 10.44GGAGNDD2032 pKa = 3.23LFYY2035 pKa = 11.24AGTAANAFTGGGGIDD2050 pKa = 3.37TVSYY2054 pKa = 10.7VSSTNGVTASLIANAVGSGGDD2075 pKa = 3.34AAGDD2079 pKa = 3.73TFSGITNLTGSAQNDD2094 pKa = 4.13TLTGLAAGGSTLNGGAGSDD2113 pKa = 3.72TLNATGDD2120 pKa = 3.55NNIFIGGAGGDD2131 pKa = 3.56TMTGNGGLNNTADD2144 pKa = 3.99YY2145 pKa = 8.94TGSGAVVVDD2154 pKa = 4.7LFFTDD2159 pKa = 3.84GTGTSGGDD2167 pKa = 3.18ATGDD2171 pKa = 3.54KK2172 pKa = 10.91LSGIQNVKK2180 pKa = 10.54GSGGNDD2186 pKa = 2.75TFFANSVANVFTGNGGTDD2204 pKa = 3.22TVSYY2208 pKa = 10.21SHH2210 pKa = 6.41STTTVIASLLAGVGGTSGDD2229 pKa = 3.71ATGDD2233 pKa = 3.45TYY2235 pKa = 11.84NAITNLTGGSGDD2247 pKa = 3.71DD2248 pKa = 3.55TLTGLLAGGSTLVGNAGNDD2267 pKa = 3.21RR2268 pKa = 11.84LIATGGNNVLMGGAGADD2285 pKa = 3.39NMSATGANNTASYY2298 pKa = 10.77AGSTNGNIIVDD2309 pKa = 4.08LTKK2312 pKa = 10.46TDD2314 pKa = 3.48GTGTSGGDD2322 pKa = 3.13AAGDD2326 pKa = 3.51ILSGIQNVIGGLGDD2340 pKa = 3.87DD2341 pKa = 5.44KK2342 pKa = 11.46FILDD2346 pKa = 4.01HH2347 pKa = 6.38NANIIDD2353 pKa = 4.39GGNGSNTVSYY2363 pKa = 10.38EE2364 pKa = 3.75HH2365 pKa = 6.86EE2366 pKa = 4.04TTINVTVDD2374 pKa = 3.23LSGTGIANGDD2384 pKa = 4.08GLFGAQDD2391 pKa = 3.71TLTNIQNAIGGGGNDD2406 pKa = 3.55IFLAGNAHH2414 pKa = 6.11NFFDD2418 pKa = 4.65GGGGVDD2424 pKa = 3.42TVSYY2428 pKa = 11.06ANNNGAHH2435 pKa = 6.39GAAIAANTAGVTVDD2449 pKa = 4.01FHH2451 pKa = 8.68GNIGSGNWAAGDD2463 pKa = 3.57TYY2465 pKa = 11.41TNIEE2469 pKa = 4.18NVIGTTGNDD2478 pKa = 3.28TFIDD2482 pKa = 3.89AGDD2485 pKa = 4.12GVHH2488 pKa = 7.04NIYY2491 pKa = 10.97NGGANGAGSNDD2502 pKa = 3.25TVSYY2506 pKa = 10.51QFSATGGALTLSIDD2520 pKa = 3.68GTHH2523 pKa = 6.83ANAGNAAGDD2532 pKa = 3.79VFTNIANLTGSDD2544 pKa = 3.65TVNSEE2549 pKa = 3.55LWGDD2553 pKa = 3.65GNANILTAMGSTNTNILHH2571 pKa = 6.24GNGGGAGTDD2580 pKa = 3.47VLDD2583 pKa = 4.56GRR2585 pKa = 11.84LGGHH2589 pKa = 5.58NTLIAGDD2596 pKa = 3.93TGSTTVDD2603 pKa = 2.95INASADD2609 pKa = 3.51PNHH2612 pKa = 7.02AGGSILGTNIDD2623 pKa = 4.02QIVGSTTGAVTVKK2636 pKa = 9.84MFNLTTALDD2645 pKa = 3.52ISSFSATATVGGPKK2659 pKa = 9.71VSHH2662 pKa = 6.31ITTLDD2667 pKa = 3.38LSGDD2671 pKa = 3.79GVGTQISISANDD2683 pKa = 3.79VIALGNNHH2691 pKa = 5.69QLSVRR2696 pKa = 11.84MDD2698 pKa = 3.69GNDD2701 pKa = 3.85SINLVLQAGEE2711 pKa = 4.02HH2712 pKa = 6.03SVLFGNGDD2720 pKa = 3.66YY2721 pKa = 11.57AFFDD2725 pKa = 3.99ANNVEE2730 pKa = 4.59LARR2733 pKa = 11.84IHH2735 pKa = 6.33QLTT2738 pKa = 3.51
Molecular weight: 267.76 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4V1QQH5|A0A4V1QQH5_9BURK Extracellular solute-binding protein OS=Oxalobacteraceae bacterium CAVE-383 OX=2478672 GN=D9O50_02575 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 9.97QPSVVRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.57RR14 pKa = 11.84THH16 pKa = 5.79GFRR19 pKa = 11.84ARR21 pKa = 11.84MATRR25 pKa = 11.84GGRR28 pKa = 11.84AVINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.58GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 9.97QPSVVRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.57RR14 pKa = 11.84THH16 pKa = 5.79GFRR19 pKa = 11.84ARR21 pKa = 11.84MATRR25 pKa = 11.84GGRR28 pKa = 11.84AVINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.58GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1228013 |
29 |
6384 |
341.0 |
37.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.389 ± 0.059 | 0.885 ± 0.014 |
5.499 ± 0.039 | 5.149 ± 0.034 |
3.701 ± 0.024 | 8.015 ± 0.053 |
2.179 ± 0.019 | 5.544 ± 0.029 |
3.948 ± 0.036 | 10.449 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.68 ± 0.022 | 3.246 ± 0.042 |
4.915 ± 0.04 | 3.948 ± 0.027 |
6.178 ± 0.052 | 5.638 ± 0.032 |
5.008 ± 0.059 | 6.916 ± 0.033 |
1.228 ± 0.015 | 2.484 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
